Figures & data
Figure 1. Workflow of ribosome profiling experiment and ribosome occupancy analysis. (A) Bergström needle biopsies from vastus lateralis muscles were obtained from healthy adults. Samples were collected from three middle-aged and two old individuals, as shown. Ribosome profiling and Total RNA libraries (RiboSeq and RNASeq, respectively) were prepared. The bioinformatic workflow for the ribosome profiling data filtered out contaminating non-coding RNA sequences, in particular rRNAs, tRNAs, snRNAs, and snoRNAs, and then aligned the RiboSeq reads to the known transcriptome. In parallel, Total RNASeq data was aligned to the transcriptome. Finally, translational efficiency (TE) was calculated for each known transcript by finding the ratio RiboSeq/RNASeq. TE was then compared between muscle from younger and older participants. (B) Overall distributions of ribosome profiling and Total RNA sequencing data. The scatterplots show, for each known transcript, the normalized level of reads from the ribosome profiling libraries in transcripts per million (TPM) in the y-axes (RiboSeqTPM) and the RNA sequencing in the x-axes (RNASeqTPM). The y-axis represents the level of ribosome occupancy and hence translation level, while the x-axis is a representation of the steady-state levels of mRNAs in the cell (the balance between mRNA transcription and mRNA degradation). Highlighted are the transcripts whose translation was significantly different between younger and older individuals: red dots represent mitochondrial transcripts, which cluster together and were decreased with age, green dots represent mRNAs showing decreasing translation in older muscle, and blue represent mRNAs showing increased translation in older muscle. Descriptions of significantly changed transcripts are found in
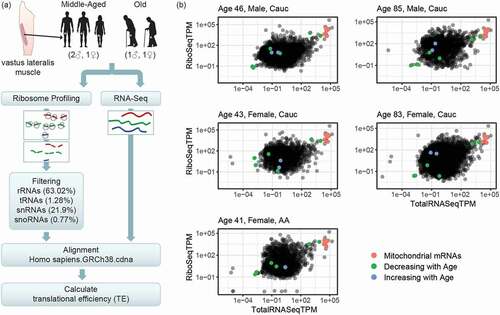
Table 1. Transcripts with translational efficiency significantly different between age groups. Negative fold changes indicate lower translation with older age and, viceversa, positive fold changes indicate higher translation with older age. Significance was tested using a Wald test with Benjamini-Hochberg adjustment
Supplemental Material
Download Zip (225 KB)Data availability statement
Raw RNASeq datasets from all samples, both RiboSeq and TotalRNASeq, are openly available in NCBI GEO at www.ncbi.nlm.nih.gov/geo with accession number GSE152558.25
