Figures & data
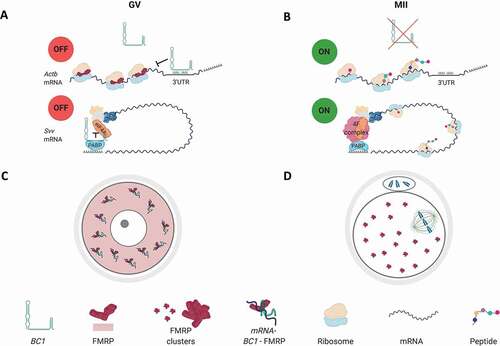
Figure 1. BC1 ncRNA is abundant in the GV oocyte with polysomal association
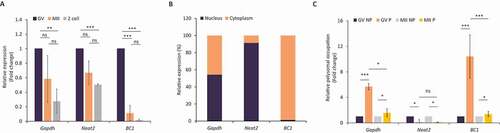
Figure 2. BC1 overexpression leads to decrease of global translation and TOP RNA reporter in the GV oocytes
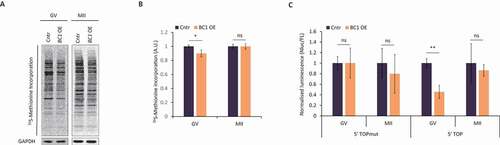
Figure 3. BC1 ncRNA influences translation of specific targets in the GV mouse oocyte
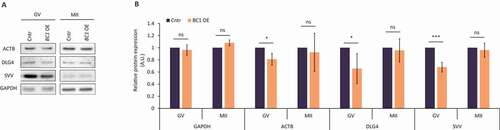
Figure 4. BC1 physically interacts with candidate mRNAs via protein interplay
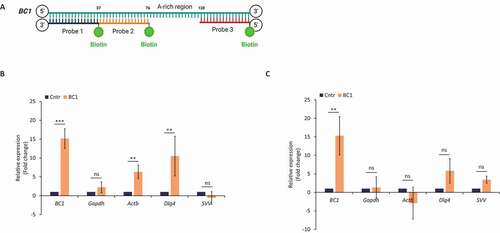
Figure 5. BC1 interacts with FMRP protein in the GV oocyte
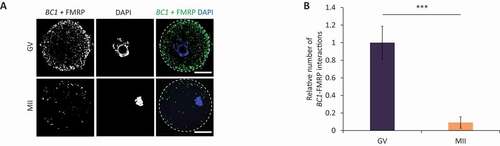
Figure 6. BC1 overexpression does not lead to clustering of FMRP and RNA in GV oocyte
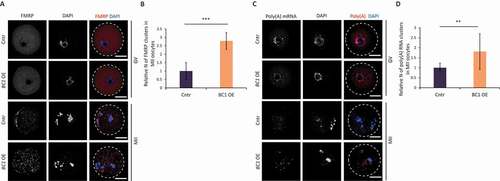
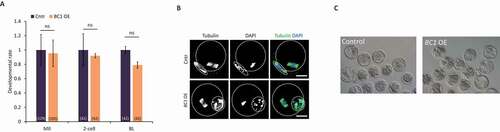
Figure 7. Overexpression of BC1 does not alter oocyte or early embryo development