Figures & data
Table 1. Proteins involved in DNA elimination in Tetrahymena
Figure 1. Tetrahymena life cycle. Each cell contains a macronucleus (MAC) and a micronucleus (MIC). In the presence of sufficient nutrients, Tetrahymena reproduces asexually by binary fission (A). However, when there is a lack of nutrients, it reproduces sexually by conjugation (B-J). To start conjugation, two cells of complementary mating types fuse (B) and their MICs undergo meiosis (C). Three of the meiotic products are degraded, while the surviving nucleus undergoes mitosis (D). The fused cells then exchange a pronucleus (E) and the pronuclei fuse to create a zygotic nucleus (F), which undergoes two rounds of mitosis (G). Two products will develop into new MACs, one will form a new MIC and the fourth product is degraded. The parental MAC is also degraded and the fused cells separate (H). Finally, the MIC divides mitotically (I), which is followed by binary fission (J). The approximate time-scale of events is indicated in hours post-mixing (hpm)
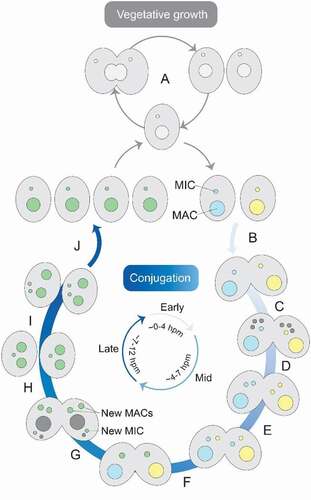
Figure 2. Programmed DNA elimination. Early scan-RNAs (scnRNAs) are produced from type-A internal eliminated sequences (IESs) and surrounding regions (A). Next, they are transported to the MAC (B) and scnRNAs complementary to the MAC genome are degraded. Meanwhile, a new MIC and MAC have been formed (C). The remaining scnRNAs are transported to the new MAC, where they recognize both type-A and – B IESs (D). The scnRNAs induce heterochromatin formation and the production of late-scnRNAs, which further spread the heterochromatin (E). Finally, the parts of the genome marked by heterochromatin are excised and the ends are ligated (F)
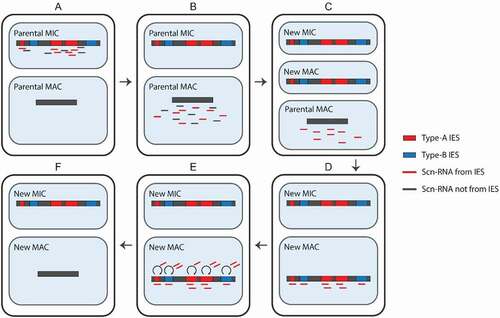
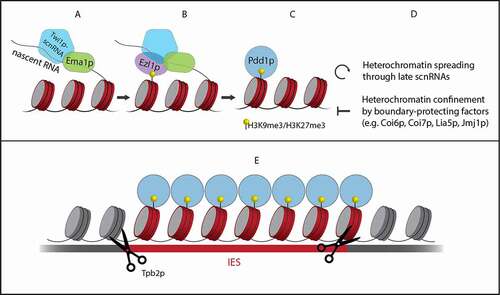
Figure 3. From Twi1p-scnRNA target search to IES elimination. Together with Ema1p, the Twi1p-scnRNA complex searches for targets in nascent transcripts (A). At matching sequences, histone H3 is methylated at lysine 9 (H3K9me3) and 27 (H3K27me3) by Ezl1p (B). Pdd1p is recruited to the heterochromatin (C). This is a requirement for the production of late scnRNAs that further spread the heterochromatin. At the same time, boundary-protecting factors confine the heterochromatin to IESs (D). Finally, Tpb2p excises IESs marked by heterochromatin (E)