Figures & data
Figure 1. Components involved with linear mRNA or circular RNA biogenesis. (a) The top panel shows a pre-mRNA transcript with four exons. Long flanking introns containing inverted repeat elements such as Alu elements or trans-acting RBP proteins bring the downstream splice site into close proximity with the upstream splice site to favour back splicing and thus circRNA production. Conversely, introns bound by ADAR1 ‘melt’ RNA secondary structures by disrupting intronic base pairing to disrupt circularization. (b) Intronic circRNA biogenesis: Linear splicing produces lariat structures that are usually debranched and hydrolysed. However, GU rich and C rich intronic motifs escape debranching to form ciRNAs (intronic circRNAs). ADAR adenosine deaminases acting on RNA, BP: Branch points. RBP: RNA binding proteins. ALU: Alu repeat elements, mRNA messenger RNA
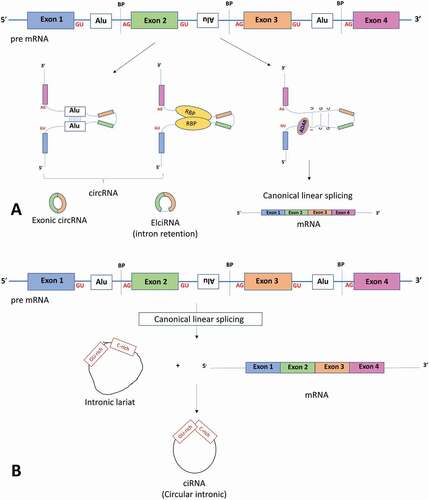
Table 1. Software for circular RNA detection and downstream applications
Figure 2. Three different strategies of identifying back-splice junction (BSJ) reads employed by circRNA detection software. (a) Pseudo reference strategy: Reads are first aligned to the reference (grey boxes) any unmapped reads (red boxes) are taken forward and aligned against pseudo references, which are all possible combinations of exon junctions. In this way, any reads that align identify the exons involved in the back-spliced junctions. (b) Split alignment strategy: Splice-aware aligners split the sequencing reads which allows direct alignment to the reference. Reads can be selected to choose the BSJ reads. (c) Kmer strategy: The actual step of read alignment is forgone. Instead, all possible kmers (short stretches of sequence) are created from the exon boundaries. Kmers are then matched to the reads. Matched kmers that are in sequence with the reference are considered linear spliced reads (top exons). Matched kmers that are out of sequence to the reference are considered circRNA, back-spliced junction reads (bottom exons)
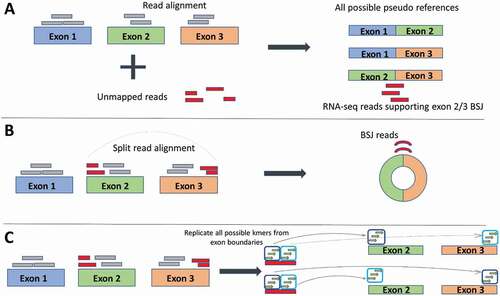
Figure 3. An overview of the various functions for circRNAs. Within the nucleus, intronic circRNAs (ciRNAs (I)) and exon and intron containing circRNAs (EIciRNAs (ii)) are involved in regulation of their parental gene. Exonic circRNAs(iii) are exported to the cytoplasm where they can either act as miRNA sponges (which also bind the Argonaute protein – an essential component of the RNA-induced silencing complex (RISC) which acts upon the targeted mRNA), RBP sponges, protein scaffolds or are translated. RBP: RNA binding protein, Ago: Argonaute protein, Alu: Alu elements
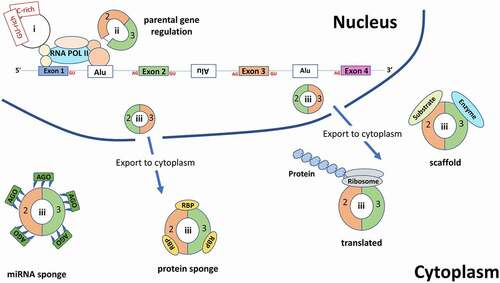
Table 2. circRNAs as potential biomarkers
Table 3. circRNAs associated with atherosclerosis
Table 4. circRNAs associated with myocardial infarction or ischaemia reperfusion injury
Table 5. circRNAs associated with heart failure/hypertrophy and cardiac fibrosis
