Figures & data
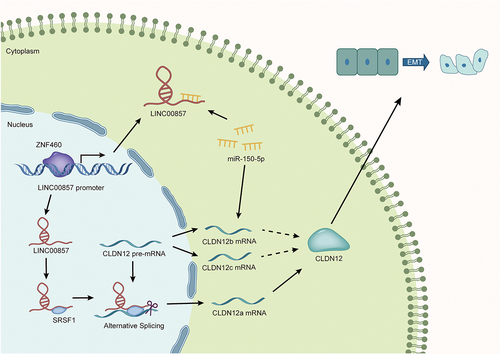
Table 1. Sequences of shRNAs
Figure 1. LINC00857 promotes malignant phenotypes of PAAD cells.
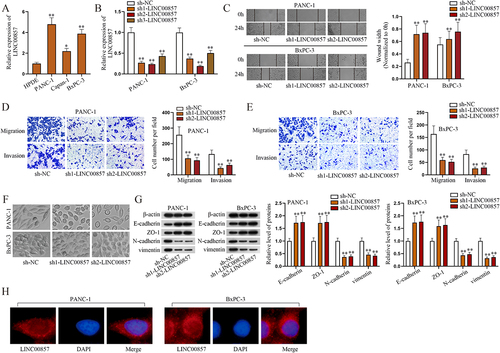
Figure 2. LINC00857 regulates the expression of CLDN12 by sponging miR-150-5p in PAAD cells.
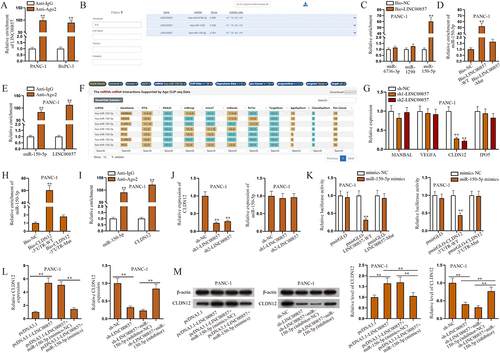
Figure 3. CLDN12 facilitates the malignant phenotypes of PAAD cells.
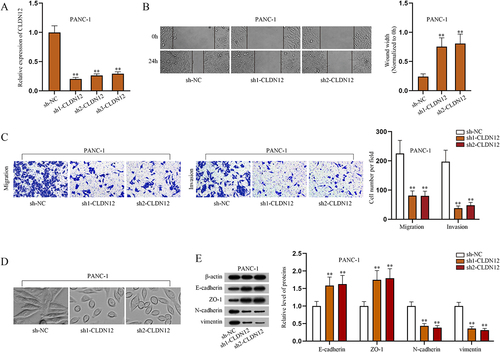
Figure 4. LINC00857 recruits SRSF1 for alternative splicing to regulate CLDN12 expression in PAAD cells.
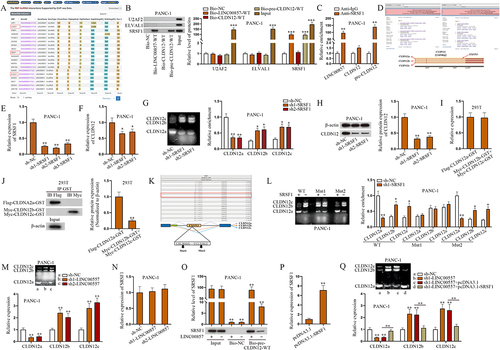
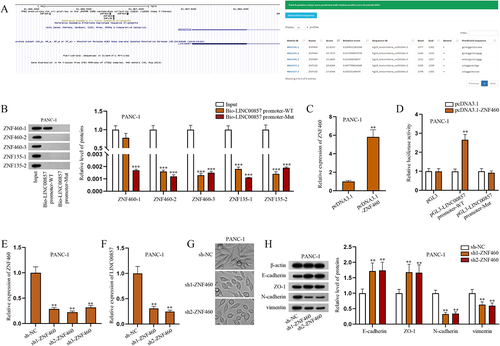
Figure 5. ZNF460 as the transcription factor of LINC00857 promoter regulates LINC00857 expression in PAAD cells.