Figures & data
Table 1. Cryo-EM structures of mitochondrial translation and ribosome rescue states
Figure 1. Schematic representation of the mitochondrial translation cycle. Mitochondrial protein synthesis follows the four steps of translation initiation, elongation, termination and ribosome recycling (for details see main text). Translation factors are depicted as follows: mtIF2, light blue; mtIF3, brown; mtEFTu, purple; mtEFTs, magenta; mtEFG1, Orange; mtRF1a, red; mtRRF, dark blue; mtEFG2, pink; MALSU1 module, turquoise; GTP and GDP are shown as red and Orange circle, respectively. Most of the depicted states were structurally resolved by cryo-EM (for PDB codes and further details, see ). The MALSU1 module composed of MALSU1, L0R8F8 and mtACP may act as an anti-association factor during ribosome recycling.
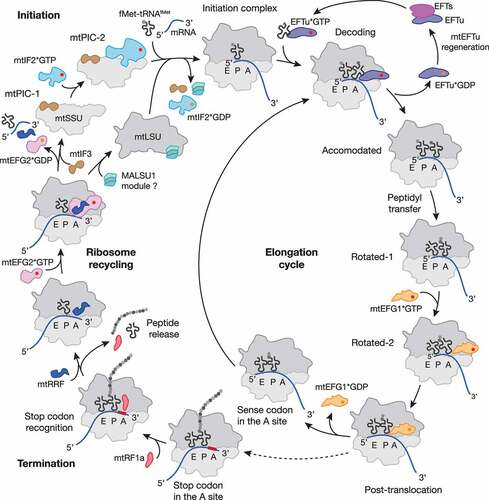
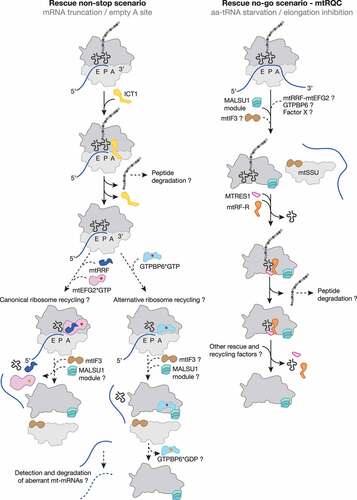
Figure 2. Mitochondrial ribosome rescue. Two pathways have recently been described involving two members of the release factor family, ICT1 (yellow) and C12ORF65 (renamed to mtRF-R, mitochondrial release factor in rescue; Orange). ICT1 targets stalled ribosome complexes with an empty A site due to truncated mRNAs (non-stop rescue pathway). Whether ribosomes follow the canonical (mtRRF, dark blue; mtEFG2, pink) or the alternative recycling pathway (GTPBP6, light blue) after ICT1-mediated peptide hydrolysis and whether aberrant mt-mRNAs can be detected and degraded are open questions. mtRF-R is required to rescue stalled complexes accumulating due to aa-tRNA starvation (no-go rescue pathway). mtRF-R acts in concert with MTRES1 (rose) on split mtLSU harbouring a peptidyl tRNA in the P site. Thus, the action of mtRF-R and MTRES1 depends on a preceding recycling event, however, it is currently unclear which factor can recycle these stalled complexes. Further translation factors are depicted as in .