Figures & data
Table 1. Subunits of archaea, yeast and human RNA polymerases
Figure 1. Human RNA polymerase III (Pol III) structure and function. (A) Important structural elements of the elongating Pol III including the bridge helix; the clamp; the trigger loop; the wall; the protrusion; the lobe; the peripheral RPC8-RPC9 stalk; the RPC3-RPC6-RPC7 heterotrimer and the RPC4-RPC5 heterodimer [Protein Data Bank (PDB) ID 7D58, 7AE1 and 7DU2]. (B) Important structural elements of the apo Pol III including the [4Fe-4S] motif in RPC6; the RPC7 stalk bridge and the RPC7 C-terminus in the active cleft and the RNA exit channel (PDB ID 7D59 and 7A6H). (C) Front view on open and closed clamp conformations in human Pol III with the closed clamp state in blue and open clamp state in grey. The corresponding values indicate the relative distance of the cleft of the two conformations. The black arrows indicate the orientation of the movement of heterotrimer and stalk during the transition from apo state to elongating state.
![Figure 1. Human RNA polymerase III (Pol III) structure and function. (A) Important structural elements of the elongating Pol III including the bridge helix; the clamp; the trigger loop; the wall; the protrusion; the lobe; the peripheral RPC8-RPC9 stalk; the RPC3-RPC6-RPC7 heterotrimer and the RPC4-RPC5 heterodimer [Protein Data Bank (PDB) ID 7D58, 7AE1 and 7DU2]. (B) Important structural elements of the apo Pol III including the [4Fe-4S] motif in RPC6; the RPC7 stalk bridge and the RPC7 C-terminus in the active cleft and the RNA exit channel (PDB ID 7D59 and 7A6H). (C) Front view on open and closed clamp conformations in human Pol III with the closed clamp state in blue and open clamp state in grey. The corresponding values indicate the relative distance of the cleft of the two conformations. The black arrows indicate the orientation of the movement of heterotrimer and stalk during the transition from apo state to elongating state.](/cms/asset/8ac72f58-b788-4f87-8af6-5caed6aa5583/krnb_a_2022293_f0001_oc.jpg)
Figure 2. The structural features of polymerase active centre in the elongating state. (A) Front view of elongating Pol III (left panel, PDB ID 7D58), Pol II (middle panel, PDB 5FLM) and Pol I (right panel, PDB ID 5M3F). The cleft distance is indicated by a dished line and the corresponding values indicate the relative distance of the cleft. (B) Close-up view of the active site of human Pol III (PDB ID 7D58), bovine Pol II (PDB ID 5FLM), bacterial Pol (PDB ID 2O5I), yeast Pol III (PDB ID 5FJ8), yeast Pol II (PDB ID 1Y1W) and yeast Pol I (PDB ID 5M3F).
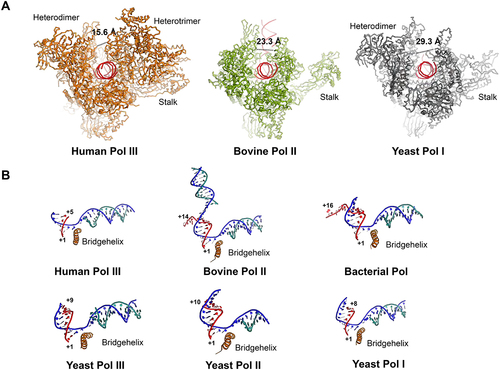
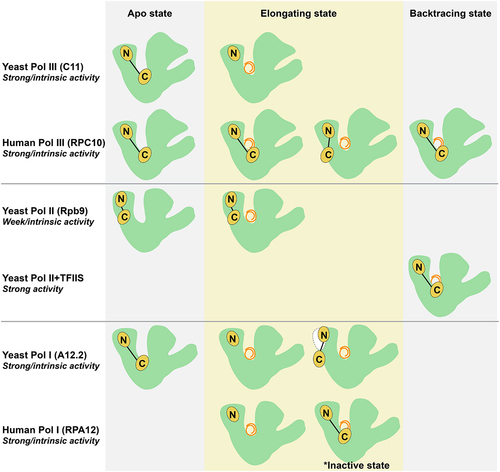
Figure 3. Schematic presentation of the RPC10 C-Zn-ribbon domain and its homologs in Pol I, Pol II and Pol III during apo, elongating and backtracking states. In the apo state, the C-Zn-ribbon of C11/RPC10/A12.2 with strong intrinsic endonuclease activity in yeast Pol III, human Pol III and yeast Pol I locate in the active site, while the C-Zn-ribbon of Rpb9 with very weak intrinsic endonuclease activity in yeast Pol II resides on the enzyme surface. In the elongating state, the RPC10 C-Zn-ribbon of human Pol III shows either ‘inside active site’ conformation, or the ‘outside funnel’ conformation, while the C11 C-Zn-ribbon of yeast Pol III disassociate from the enzyme core; the C-Zn-ribbon domain of A12.2 and RPA12 in both yeast and human Pol I are excluded from the active site, and A12.2 C-Zn-ribbon only visible on the yeast Pol I surface when A49-A34.5 dissociate from enzyme, and RPA12 C-Zn-ribbon was observed in the active site in an inactive state. In the backtracking state, the RPC10 C-Zn-ribbon of human Pol III still stays in the active site; the elongation factor TFIIS with strong endonuclease activity binds to yeast Pol II with the C-Zn-ribbon domain in the active site.