Figures & data
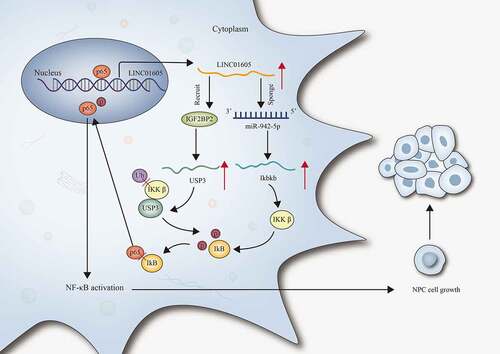
Figure 1. LINC01605 is upregulated in NPC cells.
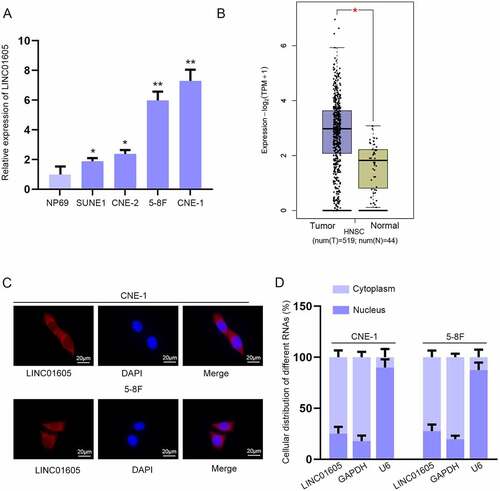
Figure 2. LINC01605 silence inhibits cell proliferation and enhances cell apoptosis in NPC. (a) Efficiency of sh-LINC01605#1/2 in CNE-1 and 5–8 F cells was evaluated by RT-qPCR. (b–d) The changes in cell viability and proliferation under the influence of sh-LINC01605 transfection were evaluated by CCK-8, EdU and colony formation assays. (e–f) Apoptosis of sh-LINC01605-transfected CNE-1 and 5–8 F cells was measured by JC-1 and TUNEL assays. One-way ANOVA was applied for statistical analysis. **P < 0.01.
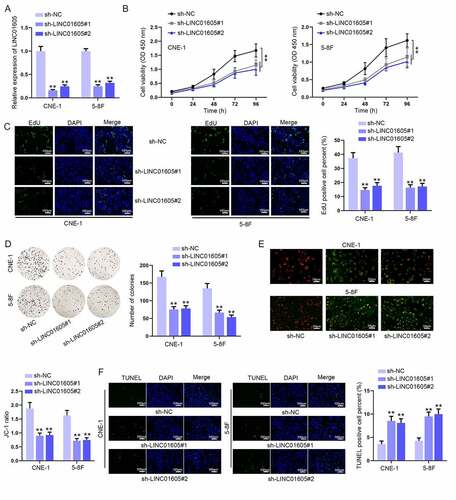
Figure 3. LINC01605 regulates Ikbkb expression to promote nuclear translocation of p65 and thereby activates the NF-κB pathway.
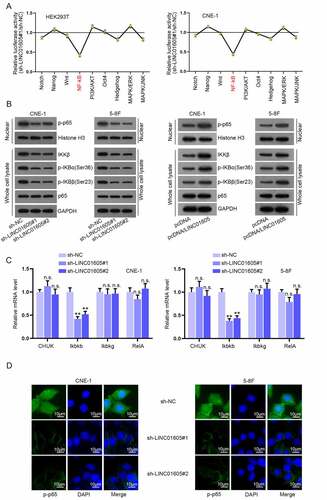
Figure 4. LINC01605 sequesters miR-942-5p to regulate Ikbkb mRNA.
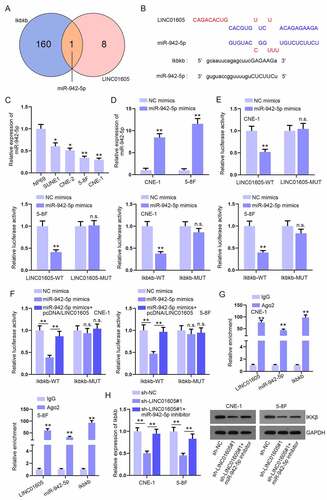
Figure 5. LINC01605 regulates USP3 to stabilize IKKβ protein.
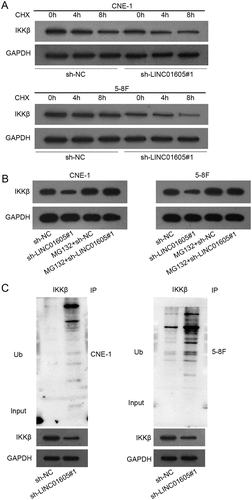
Figure 6. LINC01605 recruits IGF2BP2 to stabilize USP3 mRNA.
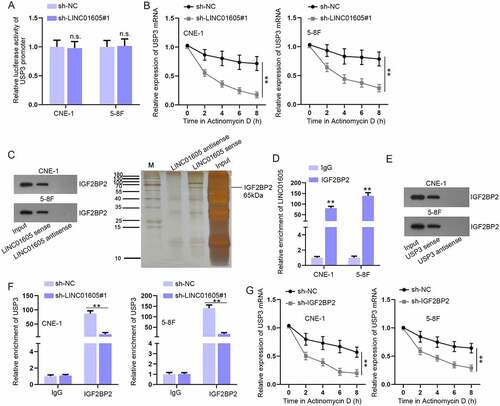
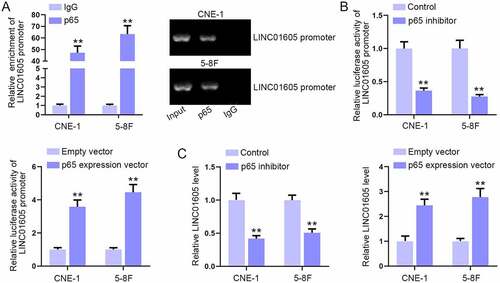
Figure 7. Overexpression of p65 enhances LINC01605 expression.