Figures & data
Figure 1. Schematic structures of human RNase MRP and RNase P. (a) Secondary structure of the RNase MRP and P RNAs with the relative positions of the protein subunits based on intermolecular interaction data. It is unclear whether Rpp14, Rpp21 and hPop4 (depicted in grey) stably bind to the RNase MRP complex. Figure adapted from Welting et al [Citation1]. (b). RNase MRP mutations used in this study. (c) The S1m aptamer (blue) is fused to either the 5’- and 3’-end of the MRP RNA (black) or inserted internally (int) between nucleotides 158 and 159. In case of the RNase P RNA the S1m-aptamer was inserted internally between nucleotides 209 and 210.
![Figure 1. Schematic structures of human RNase MRP and RNase P. (a) Secondary structure of the RNase MRP and P RNAs with the relative positions of the protein subunits based on intermolecular interaction data. It is unclear whether Rpp14, Rpp21 and hPop4 (depicted in grey) stably bind to the RNase MRP complex. Figure adapted from Welting et al [Citation1]. (b). RNase MRP mutations used in this study. (c) The S1m aptamer (blue) is fused to either the 5’- and 3’-end of the MRP RNA (black) or inserted internally (int) between nucleotides 158 and 159. In case of the RNase P RNA the S1m-aptamer was inserted internally between nucleotides 209 and 210.](/cms/asset/6572f005-b669-4df0-a6c4-2a32a9750ad4/krnb_a_2027659_f0001_oc.jpg)
Figure 2. Ribonuclease activity of S1m-MRP and S1m-P complexes. A radiolabeled viperin mRNA fragment and pre-tRNA were incubated for 1.5 hrs with purified S1m-MRP and S1m-P complexes as indicated. The input material and the substrate incubated for 1.5 hrs in the absence of purified material are indicated by t = 0 and t = 1.5 hrs, respectively. Mock refers to the incubation of the substrates with material isolated from a cell lysate of untransfected cells using the same procedure as for the lysates from S1m-RNA expressing cells. Immunoprecipitated material (IP, with an antibody to Rpp20) was used as a control for affinity-purified RNase MRP and RNase P. NRS: normal rabbit serum. * and ** mark the positions of the substrate RNAs and their major cleavage products, respectively.
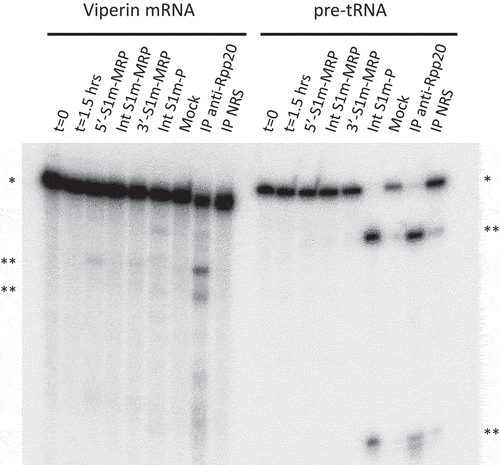
Figure 3. Expression levels of S1m-tagged MRP RNA mutants. HEK293T cells were transfected with S1m-tagged wild type (WT) and mutant MRP RNA constructs (indicated by the nucleotide changes) and the expression levels after 48 hours were determined by northern blot hybridization. Expression levels of (a) 5’-S1m-MRP RNAs and (b) 3’-S1m-MRP RNAs. Levels were normalized based on U3 snoRNA levels and the level of wildtype S1m-MRP was set to 100%. A t-test was performed to determine significance * = P < 0.05, ** = P < 0.01.
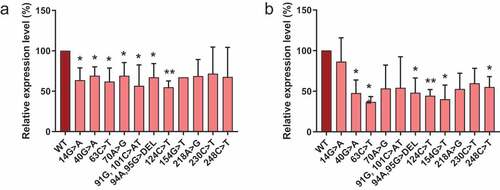
Figure 4. Association of Rpp25 with 3’-S1m-MRP RNAs. 3’-S1m-MRP RNAs (mutants indicated by nucleotide substitutions) were isolated from transfected HEK293T cells using streptavidin beads. Input, non-bound and bound material were analysed by Western blotting using a polyclonal anti-Rpp25 antibody. Equivalent amounts of input and non-bound fractions were loaded. The relative amount of bound material analysed was based on the amount of bound S1m-MRP quantified by northern blotting. Mock refers to material isolated from a cell lysate of untransfected cells.
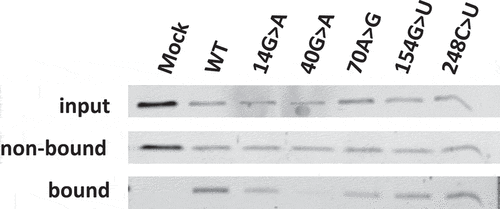
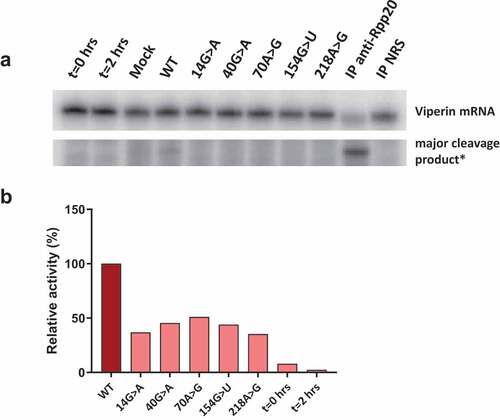
Figure 5. Catalytic activity of wild type and mutant RNase MRP complexes. 3’-S1m-MRP RNA-containing complexes were isolated from transfected cell lysates and incubated with the radiolabeled viperin mRNA fragment. Reaction products were analysed by denaturing gel electrophoresis and autoradiography. A) Substrate RNA and major cleavage products. *Note that the results are from the same gel, but with different exposure times (longer exposure for detection of cleavage product). Material immunoprecipitated (IP) with anti-Rpp20 antibodies and with antibodies from normal rabbit serum (NRS) was used as control. B) Relative cleavage activity observed for the 3’-S1m-MRP complexes. Activity of the wild type (WT) 3’-S1m-MRP complex was set to 100%. Lanes marked t = 0 hrs and t = 2 hrs represent the viperin mRNA substrate incubated for 0 and 2 hours in the absence of isolated material.