Figures & data
Figure 1. RNA chaperone functions in the bacterial cell. RNA chaperones facilitate proper RNA folding, remodel existing states, assist in matchmaking between regulatory RNAs and their targets, protect bound RNA from ribonucleases, and by the interaction with the ribosome-binding site (RBS) activate or inactivate translation.
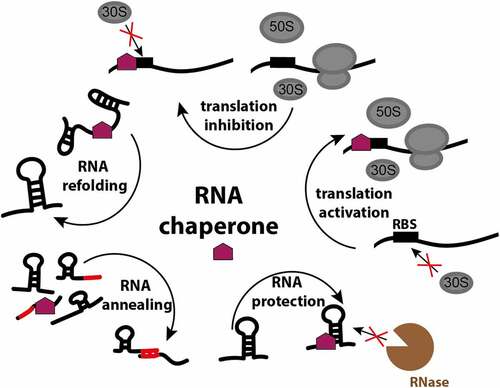
Table 1. Distribution of RNA chaperones and riboregulators in Bacteria, and representative species for the groups. Hfq, ProQ and CsrA have not been described in all species within these groups
Figure 2. Structures of E. coli Hfq-RNA complexes. The hexameric architecture of Hfq (grey) exposes three RNA binding surfaces: proximal face (characterised by the N-terminal α-helix, shown in orange), rim and distal face. Three views of the Hfq-RNA complexes are shown, with the proximal face (left), the rim (centre), and the distal face (right) of Hfq in the foreground. (A) class I sRNAs interact with Hfq through the proximal face and the rim. Here, the E. coli Hfq-RydC complex is used as a model of this mode of binding (PDB: 4V2S). (B) Class II sRNAs bind to the proximal and distal faces of Hfq. E. coli Hfq-3’ETSleuZ have been isolated from a ternary complex with PNPase to depict this mode of binding (PDB: 7OGM).
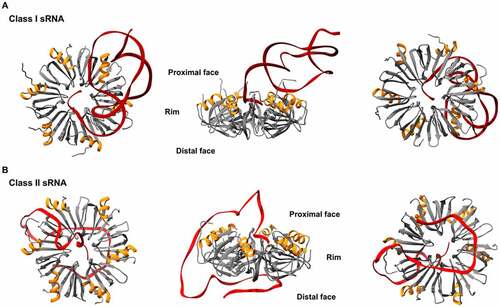
Figure 3. Structure and model for RNA binding of the N-terminal domain of E. coli ProQ. (A) N-terminal domain (NTD) of E. coli ProQ, which has the fold of the FinO domain (PDB: 5NB9). (B) ProQ NTD can be divided into two faces: a concave face, and a convex face. The concave face is RNA binding, as it presents patches of positively (in blue) and negatively (in red) charged residues. Positively charged patches on the concave face are conserved in other FinO proteins. The convex face also presents smaller areas of both negatively and positively charged residues. (C) Model of RNA binding to the NTD of ProQ. Due to the larger patches of positive residues, the concave face of the finO-domain of ProQ is expected to be mainly involved in the binding to the Rho-independent terminator hairpin at the 3’. The convex face could interact with single stranded regions nearby the double stranded hairpin [Citation61]. (D) Structure of the CTD of E. coli ProQ (PDB: 5NBB). The C-terminal domain has a Tudor-domain like structure. On the right panel, the three residues G189, T204 and G220 (in E. coli) that have been found as important for ProQ function in gene expression regulation are highlighted in yellow. These residues are exposed on the surface of the domain and are conserved in homologous proteins [Citation62]. (E) Distribution of the CTD of ProQ. The CTD is less conserved compared to the FinO-like NTD, as it is found only in γ-proteobacterial ProQ. In E. coli and Salmonella ProQ, the CTD is connected to the N-terminal FinO-like domain via a flexible 50 amino acids linker. The CTD is not found in the FinO protein, and it is expected to expand the ProQ interactome, by conferring it with ability to bind different RNA substrates [Citation66].
![Figure 3. Structure and model for RNA binding of the N-terminal domain of E. coli ProQ. (A) N-terminal domain (NTD) of E. coli ProQ, which has the fold of the FinO domain (PDB: 5NB9). (B) ProQ NTD can be divided into two faces: a concave face, and a convex face. The concave face is RNA binding, as it presents patches of positively (in blue) and negatively (in red) charged residues. Positively charged patches on the concave face are conserved in other FinO proteins. The convex face also presents smaller areas of both negatively and positively charged residues. (C) Model of RNA binding to the NTD of ProQ. Due to the larger patches of positive residues, the concave face of the finO-domain of ProQ is expected to be mainly involved in the binding to the Rho-independent terminator hairpin at the 3’. The convex face could interact with single stranded regions nearby the double stranded hairpin [Citation61]. (D) Structure of the CTD of E. coli ProQ (PDB: 5NBB). The C-terminal domain has a Tudor-domain like structure. On the right panel, the three residues G189, T204 and G220 (in E. coli) that have been found as important for ProQ function in gene expression regulation are highlighted in yellow. These residues are exposed on the surface of the domain and are conserved in homologous proteins [Citation62]. (E) Distribution of the CTD of ProQ. The CTD is less conserved compared to the FinO-like NTD, as it is found only in γ-proteobacterial ProQ. In E. coli and Salmonella ProQ, the CTD is connected to the N-terminal FinO-like domain via a flexible 50 amino acids linker. The CTD is not found in the FinO protein, and it is expected to expand the ProQ interactome, by conferring it with ability to bind different RNA substrates [Citation66].](/cms/asset/3b5c87b9-d5fe-4749-8085-b1de1b8282d6/krnb_a_2048565_f0003_oc.jpg)
Figure 4. E. coli CsrA structure and regulatory circuit. (A) CsrA forms a dimer (green and brown) with two surfaces for RNA (red) binding. (B) Schematic of CsrA regulation. The expression of csrA is regulated on transcription, translation and protein levels: CsrA can regulate its own transcription and translation. It also can be sequestered by sRNAs CsrB/C, GadY and McaS. Moreover, CsrA activity is blocked by proteins CesT in E. coli and FliW in flagellated bacteria.
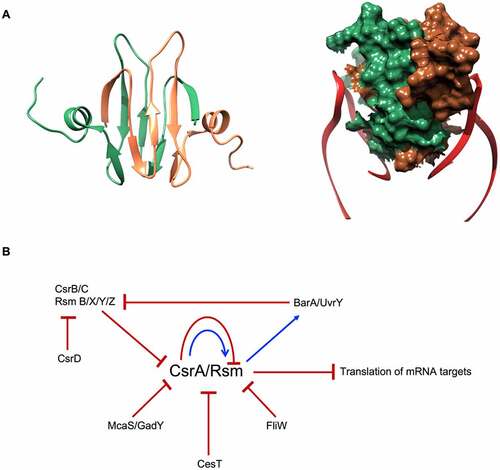
Figure 5. The C-terminal tails of Hfq: conservation and possible roles. A) Hfq sequences in different bacterial species. Residue numbering is according to E. coli Hfq. Amino acids are coloured according to their polarity (yellow – non-polar, green – polar, red – acidic, blue – basic). Alignment was performed using the MAFFT server at EBI [Citation147]. B) Predicted roles of the Hfq CTD that aid the recycling of the RNA chaperone. Upon RNA duplex formation, the CTD of Hfq facilitates the rapid release of the dsRNA (left). Hfq CTD-rim interactions also increase the selectivity of RNA binding. Through their binding to the proximal and distal faces of Hfq, Class II sRNAs are generally less susceptible to the action of Hfq C-terminal tails and can outcompete the more sensitive Class I sRNAs (right).
![Figure 5. The C-terminal tails of Hfq: conservation and possible roles. A) Hfq sequences in different bacterial species. Residue numbering is according to E. coli Hfq. Amino acids are coloured according to their polarity (yellow – non-polar, green – polar, red – acidic, blue – basic). Alignment was performed using the MAFFT server at EBI [Citation147]. B) Predicted roles of the Hfq CTD that aid the recycling of the RNA chaperone. Upon RNA duplex formation, the CTD of Hfq facilitates the rapid release of the dsRNA (left). Hfq CTD-rim interactions also increase the selectivity of RNA binding. Through their binding to the proximal and distal faces of Hfq, Class II sRNAs are generally less susceptible to the action of Hfq C-terminal tails and can outcompete the more sensitive Class I sRNAs (right).](/cms/asset/89059117-45b9-4ea5-99e1-2b72a63421e7/krnb_a_2048565_f0005_oc.jpg)
Figure 6. Hfq-Crc cooperation in Pseudomonas aeruginosa. When succinate, the preferred carbon source, is available, Hfq and Crc bind to the mRNA target, amiE, masking the ribosome-binding site and preventing its translation [Citation119,Citation120]. The distal face of captures ARN-rich repeat motifs near the RBS, where A- and R-bases occupy basic pockets on the Hfq distal side, leaving the RNA backbone and N-bases exposed to Crc. In contrast, when succinate levels are low, CrcZ, an sRNA rich in ARN-motif, is expressed to sequester Hfq away from substrate RNAs, such as amiE, allowing ribosomes to bind and begin translation [Citation119].
![Figure 6. Hfq-Crc cooperation in Pseudomonas aeruginosa. When succinate, the preferred carbon source, is available, Hfq and Crc bind to the mRNA target, amiE, masking the ribosome-binding site and preventing its translation [Citation119,Citation120]. The distal face of captures ARN-rich repeat motifs near the RBS, where A- and R-bases occupy basic pockets on the Hfq distal side, leaving the RNA backbone and N-bases exposed to Crc. In contrast, when succinate levels are low, CrcZ, an sRNA rich in ARN-motif, is expressed to sequester Hfq away from substrate RNAs, such as amiE, allowing ribosomes to bind and begin translation [Citation119].](/cms/asset/bc79fd46-46c1-485c-a399-5349e03646c9/krnb_a_2048565_f0006_oc.jpg)
Figure 7. Model of Hfq-RNase E interaction in E. coli. The interaction between Hfq and RNase E is RNA-mediated. The ternary complex formed by Hfq-sRNA-mRNA interacts with two RNA binding regions on the C-terminal domain of RNase E, namely RNA-binding domain (RBD) and second arginine-rich region (AR2) labelled in red [Citation37]. The CTD of RNase E holds the ternary complex in positions and facilitate the delivery of the mRNA target to its catalytic core. Additional components of the E. coli degradosome (i.e. RhlB, enolase and PNPase) are also shown.
![Figure 7. Model of Hfq-RNase E interaction in E. coli. The interaction between Hfq and RNase E is RNA-mediated. The ternary complex formed by Hfq-sRNA-mRNA interacts with two RNA binding regions on the C-terminal domain of RNase E, namely RNA-binding domain (RBD) and second arginine-rich region (AR2) labelled in red [Citation37]. The CTD of RNase E holds the ternary complex in positions and facilitate the delivery of the mRNA target to its catalytic core. Additional components of the E. coli degradosome (i.e. RhlB, enolase and PNPase) are also shown.](/cms/asset/cac0cf05-30b0-4e5f-ac35-c04a02f2fdbe/krnb_a_2048565_f0007_oc.jpg)
Figure 8. Hfq forms a protective complex with the exoribonuclease PNPase. In E. coli, PNPase (teal) can act as a conditional chaperone when bound with sRNA and Hfq (Orange; right panel) [Citation131]. In the absence of Hfq, PNPase can either degrade RNA (left panel) or add heterogeneous tails to existing RNA molecules (bottom panel).
![Figure 8. Hfq forms a protective complex with the exoribonuclease PNPase. In E. coli, PNPase (teal) can act as a conditional chaperone when bound with sRNA and Hfq (Orange; right panel) [Citation131]. In the absence of Hfq, PNPase can either degrade RNA (left panel) or add heterogeneous tails to existing RNA molecules (bottom panel).](/cms/asset/4343c920-40c6-450e-8051-f45cb2181982/krnb_a_2048565_f0008_oc.jpg)
