Figures & data
Figure 1. Small RNA molecules in serum of five muscular dystrophies. A substantial proportion of the circulating serum small RNAs content from each of the muscular dystrophies, A) DMD, B) DM1, C) DM2, D) FSHD1 and E) LGMD R1 calpain3-related, was determined. F) The percentages of distinct small RNA molecules population in each type of muscular dystrophy.
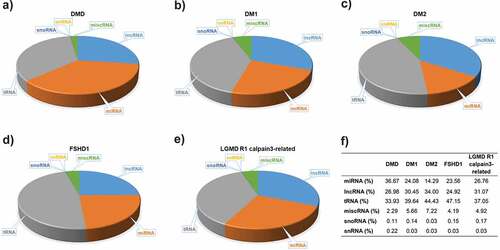
Figure 2. Differentially expressed miRNAs (DEmiRNAs) between control and patient of each muscular dystrophies. Heatmaps of the top DEmiRNAs based on p-value for each of the five diseases A) DMD, B) DM1, C) DM2, D) FSHD1 and E) LGMD R1 calpain3-related. The controls group (healthy individuals) is shown in grey colour and the muscular dystrophy patients’ group is shown in black colour. The colour key panel shows the Z-score values calculated for each miRNA, by subtracting the row-mean and then dividing by the standard deviation. Z-scores describe the expression of each miRNA in relation to the mean. Overexpressed miRNAs are shown in red, under-expressed miRNAs in blue. White colour indicates expression change close to 0. Hierarchical clustering was performed for samples and miRNAs.
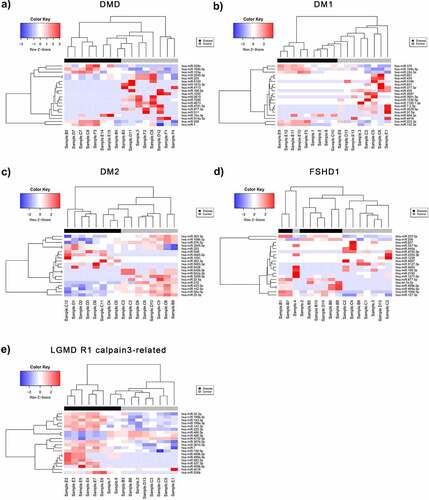
Table 1. ROC analysis for the polled miRNAs for each of the five muscular dystrophies
Figure 3. Venn diagram showing the number of unique and common predicted miRNA target genes annotated with MeSH disease terms relevant to muscular dystrophies. A) Gene targets predicted from the significantly DEmiRNAs (p-value <0.05) per disease. B) Gene targets predicted from the significantly DEmiRNAs after adjusting for multiple testing (adjusted p-value <0.05). C) Disease specific Medical Subject Headings associated with the gene targets predicted from the miR set identified after p-value adjustment.
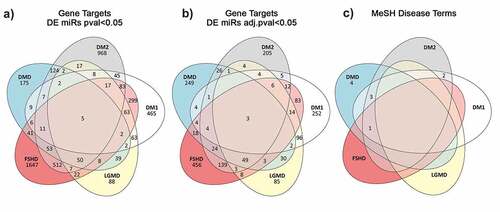
Figure 4. Enriched biological processes based on the predicted target genes for each disease. Bar dimensions represent log (Fold Enrichment score) obtained for each process. Bar colour represents the adjusted p-value with blue being of high significance and yellow close to the 0.05 adj. p-value threshold. The left panel are the processes related to muscle and neuronal activity while the right panel shows other identified processes.

Figure 5. Flow Chart of the classification process using a SVM classifier and edgeR as the feature (miRNA) selection methods. Step 1 – Describes the dataset with two classes of patients from five different types of muscular dystrophy and their control samples. Step 2 – Denotes the selection of a set of miRNAs to be used during the leave-one-out classification (LOOCV) process. Step 3 – The LOOCV is initiated by extracting one sample from the dataset. Step 4 – edgeR is used to perform differential expression analysis on the remaining samples (this avoids overfitting). X number of top significant miRNAs are used for the next. Step 5 – A SVM model is trained using the data and features for the specific iteration. Step 6 – the LOOCV process (Steps 3–5) is repeated for every sample (N = #of muscular dystrophy patients) and statistics recorded. Step 7 – Steps 2–5 are repeated for every value of X.
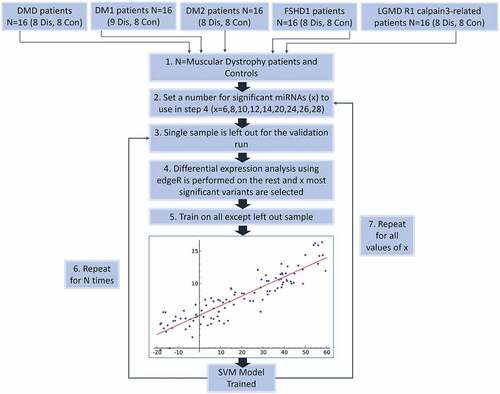
Table 2. Source edge lists databases for miRNA – gene target predictions
