Figures & data
Figure 1. Schematic structure of the snR30 RNP. This representation displays the secondary structure of snR30 comprised of the 5ʹ, internal and 3ʹ hairpins as well as the associated H/ACA proteins (Cbf5 – orange, Nop10 - blue, Gar1 – yellow, and Nhp2 - green). The snR30 RNP also interacts with the ribosome assembly factor Utp23 (purple). The m1 and m2 motifs in the 3ʹ-hairpin base-pair with expansion segment 6 (ES6) of 18S rRNA (rm1 and rm2 motifs, red). Nucleotide sequences for the H and ACA boxes as well as the m1, m2 (snR30) and rm1, and rm2 (18S rRNA) sequences are displayed.
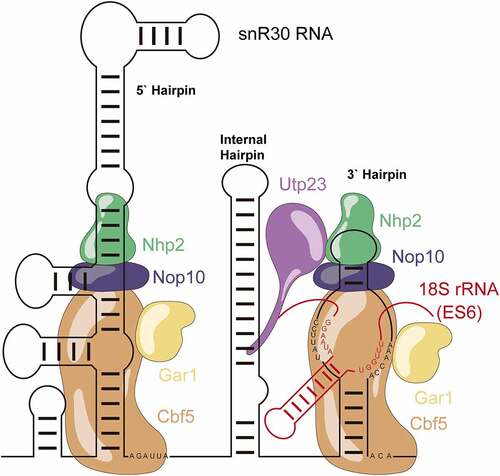
Table 1. Summary of plasmids, primers and DNA products for in vitro transcription
Table 2. Affinity of snR30 full-length and truncations binding to the H/ACA proteins Cbf5, Nop10, Gar1 and Nhp2. Dissociation constants (KD) were determined by nitrocellulose filtration () and are reported with the standard deviation
Figure 2. Binding of snR30 to the H/ACA proteins Cbf5-Nop10-Gar1-Nhp2 (CNGP). Nitrocellulose filtration assays were conducted using snR30 full-length and truncations while titrating Cbf5-Nop10-Gar1-Nhp2 protein complex. Fitting to a hyperbolic equation yielded the dissociation constants and binding amplitudes (summarized in ).
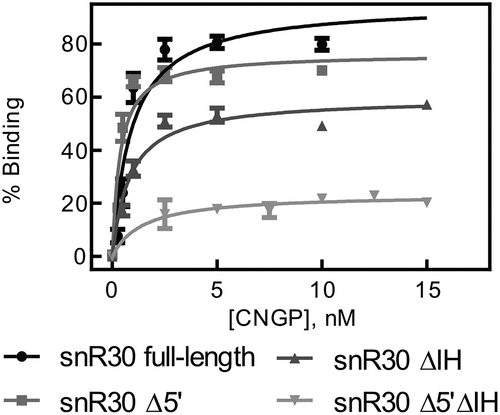
Table 3. Affinity of Utp23 for snR30 full-length and truncations. The dissociation constants for were measured by nitrocellulose filtration () and are reported with the standard deviation
Figure 3. Determining the affinity of Utp23 for snR30. Utp23 was titrated against full-length snR30 and truncations thereof as well as the modification H/ACA snoRNA snR81 as control. Hyperbolic fitting provided the dissociation constants (see ).
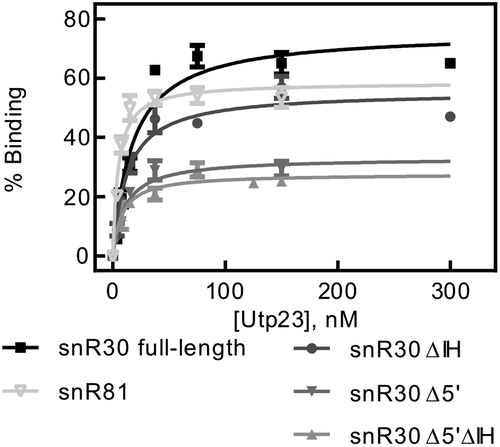
Figure 4. 18S rRNA fragments comprising and surrounding expansion segment 6 (ES6). A Diagram of the 18S rRNA and surrounding regions indicating the various rRNA constructs used in this study. B Secondary structure of the ES6 and surrounding regions as observed in the mature 80S ribosome. C Binding of snR30 full-length complexed with Cbf5-Nop10-Gar1-Nhp2 to 18S rRNA fragments containing the rm1-rm2 motif as determined by nitrocellulose filtration. D Interaction of the full-length snR30 RNP with rRNA constructs lacking the rm1-rm2 motif as well as tRNA as control. E and F Similar nitrocellulose filtration assays using snR30 RNP reconstituted with snR30 Δ5ʹ. All dissociation constants were determined by hyperbolic fitting (smooth lines) and are summarized in .
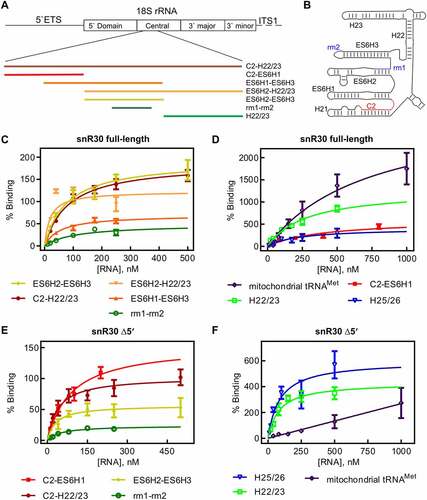
Table 4. Affinity of the snR30 RNP complex binding to fragments of ES6 of 18S rRNA. The snRN30 RNP complex was titrated with rRNA fragments and binding was quantified by nitrocellulose filtration (). Dissociation constants are stated with the standard deviation
Table 5. Affinity of Utp23 binding to 18S rRNA fragments. The dissociation constants and binding amplitudes were determined by nitrocellulose filtration () and are summarized together with the standard deviation
Figure 5. Nitrocellulose filtration assays to quantify binding of Utp23 to 18S rRNA fragments. A Utp23 binding to 18S rRNA fragments harbouring the rm1-rm2 motif. B Binding curves for Utp23 interaction with 18S rRNA which lacks the rm1-rm2 motif as well as two control RNAs.
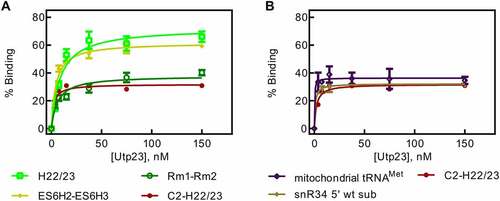
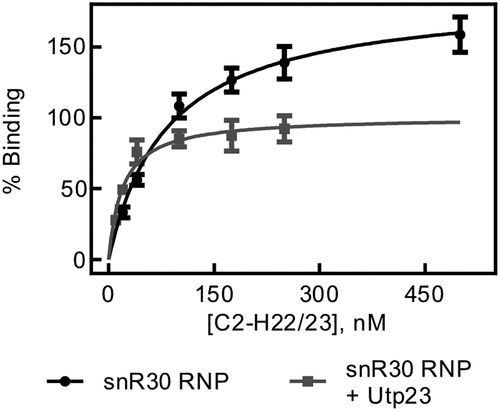
Figure 6. rRNA binding by snR30 RNP in presence of Utp23. Nitrocellulose filtration was used to quantify binding of the snR30 RNP (5 nM) to the region of 18S rRNA comprising the ES6 and flanking helices (C2-H22/23, )) in the presence of 5 nM Upt23. For comparison, binding of snR30 RNP to this rRNA fragment in the absence of Utp23 is also shown (same as in ). Hyperbolic fitting determined that snR30 RNP binds rRNA in the presence of Utp23 with a dissociation constant of 20 ± 5 nM and an amplitude of 101 ± 6%.
Supplemental Material
Download PDF (914.9 KB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
