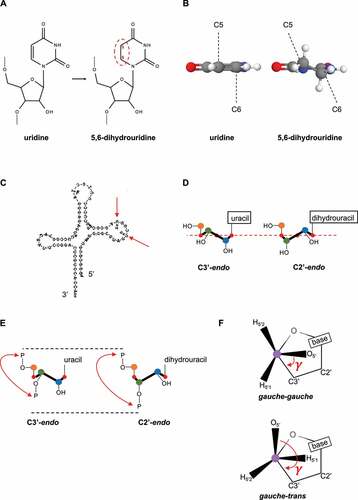
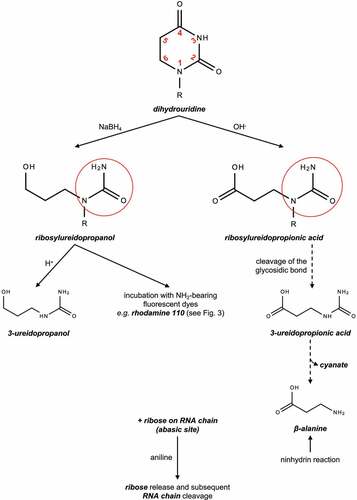
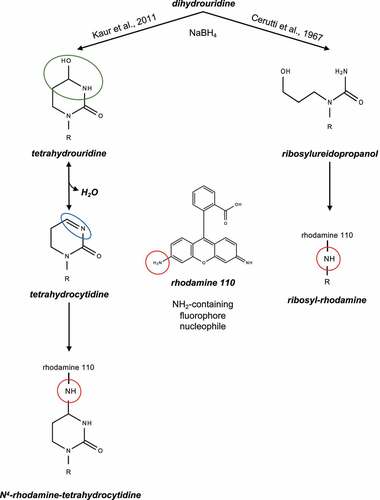
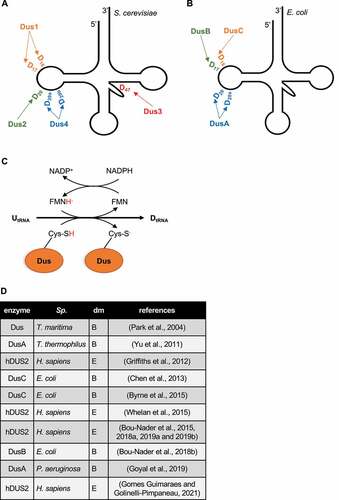
Figures & data
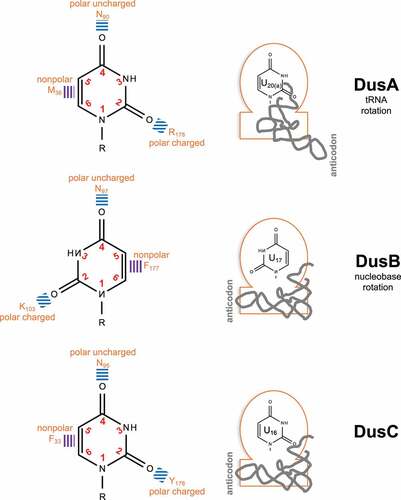
Table 1. Seminal studies on optical and structural properties of dihydro-uracil/-uridine
Table 2. Chemical reactions and techniques specifically applicable to D