Figures & data
Figure 1. Secondary structure of the 18S rRNA. (A) 18S rRNA with the 5’, central (C), 3’ major (3’M) and 3’ minor (3’m) domains indicated in different colours. All known and predicted snoRNA binding sites [Citation13,Citation19,Citation22,Citation23,Citation114,Citation120,Citation121,Citation129,Citation135] are indicated by black/grey (C/D box) or red/pink (H/ACA box) lines, and modification sites are indicated by circles. In the case of the U3 snoRNA, only the hybridization sites observed in cryo-EM structures are indicated in solid lines, while potential additional hybridization sites suggested by biochemical experiments are indicated as dashed lines. The binding regions of RNA helicases, determined by CRAC, and of the Fal1 helicase cofactor Sgd1 [Citation64,Citation135,Citation178,Citation232,Citation238] are indicated. (B) Successive folding of the 18S rRNA [Citation23]. Already folded rRNA elements are displayed in bright colours (unfolded regions in faint colours).
![Figure 1. Secondary structure of the 18S rRNA. (A) 18S rRNA with the 5’, central (C), 3’ major (3’M) and 3’ minor (3’m) domains indicated in different colours. All known and predicted snoRNA binding sites [Citation13,Citation19,Citation22,Citation23,Citation114,Citation120,Citation121,Citation129,Citation135] are indicated by black/grey (C/D box) or red/pink (H/ACA box) lines, and modification sites are indicated by circles. In the case of the U3 snoRNA, only the hybridization sites observed in cryo-EM structures are indicated in solid lines, while potential additional hybridization sites suggested by biochemical experiments are indicated as dashed lines. The binding regions of RNA helicases, determined by CRAC, and of the Fal1 helicase cofactor Sgd1 [Citation64,Citation135,Citation178,Citation232,Citation238] are indicated. (B) Successive folding of the 18S rRNA [Citation23]. Already folded rRNA elements are displayed in bright colours (unfolded regions in faint colours).](/cms/asset/5b2d030f-1dd9-42be-9d0d-02f7d519ddc0/krnb_a_2079890_f0001_oc.jpg)
Figure 2. Secondary structure of the 25S and 5.8S rRNAs. (A) 25S rRNA with domains 0 to VI indicated in different colours. All known and predicted snoRNA binding sites [Citation114,Citation120,Citation121] are indicated by black/grey (C/D box) or red/pink (H/ACA box) lines, and modification sites are indicated by circles. The binding regions of RNA helicases, determined by CRAC [Citation64,Citation69,Citation232,Citation238,Citation265], are indicated. Additionally, the binding sites of Npa1, an interaction partner of RNA helicase Dbp6 [Citation52], are indicated. (B) Successive folding of the 25S rRNA [Citation54,Citation61]. Already folded rRNA elements are displayed in bright colours (unfolded regions in faint colours).
![Figure 2. Secondary structure of the 25S and 5.8S rRNAs. (A) 25S rRNA with domains 0 to VI indicated in different colours. All known and predicted snoRNA binding sites [Citation114,Citation120,Citation121] are indicated by black/grey (C/D box) or red/pink (H/ACA box) lines, and modification sites are indicated by circles. The binding regions of RNA helicases, determined by CRAC [Citation64,Citation69,Citation232,Citation238,Citation265], are indicated. Additionally, the binding sites of Npa1, an interaction partner of RNA helicase Dbp6 [Citation52], are indicated. (B) Successive folding of the 25S rRNA [Citation54,Citation61]. Already folded rRNA elements are displayed in bright colours (unfolded regions in faint colours).](/cms/asset/3fd3d2a9-3f69-4ae3-914e-d506f3feb1f5/krnb_a_2079890_f0002_oc.jpg)
Figure 3. rRNA folding steps shaping the evolving 90S/pre-40S and pre-60S particles. 18S (A) or 5.8S, 25S, and 5S (B) (pre-) rRNA domains are colour coded and the consecutive RNA shaping events illustrated schematically (left panels) or using existing cryo-EM structures of ribosomal maturation intermediates (right (A) and middle panels (B)). Association/dissociation of RNA helicases at distinct pre-60S maturation stages is indicated using the colour codes of their potential rRNA target domains ((B), right panel). PDB codes for 90S/pre-40S structures (A) (from top to bottom): 6ZQA (state A), 6ZQB (state B), 6ZQC (state C/pre-A1), 6LQS (state D/post-A1), 6ZQE (state Dis-A), 6ZQG (state Dis-C), 4v88 (mature 40S). PDB codes for pre-60S structures (B) (from top to bottom): 6EM3 (state A), 6EM4 (state B), 6EM1 (state C), 6ELZ (state D/E), 6YLX (state NE – Nop53 early), 3JCT (state Nog2/F), 6YLG (state LN/Rix1-Rea1), 4v88 (mature 60S).
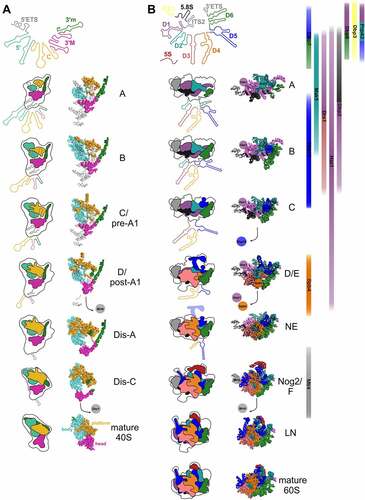
Table 1. RNA helicases in ribosome biogenesis and their presumed functions
Figure 4. Cryo-EM structures of helicases Dhr1 and Has1 bound to their pre-ribosomal substrate particles. (A) Dhr1 (red) bound to 90S (left panel, PDB: 6ZQD) or pre-40S (right panel, PDB: 6ZQG) particles. The assembly factors Utp14 (cyan) and Pno1 (blue) in proximity to the Dhr1 C-terminus, as well as the U3 snoRNA (black) and 25S rRNA H1 (Orange, right panel) base-pairing with U3 box A are depicted. (B) Has1 (red) binds to pre-60S intermediates (PDB: 5Z3G) on top of the ITS2-containing foot structure close to Nsa3. The two RecA-like domains interact with H16 (purple) of 25S rRNA domain I. The Has1 25S rRNA crosslink site at H21/H22 (Orange) and further proximal assembly factors and -r-proteins are indicated.
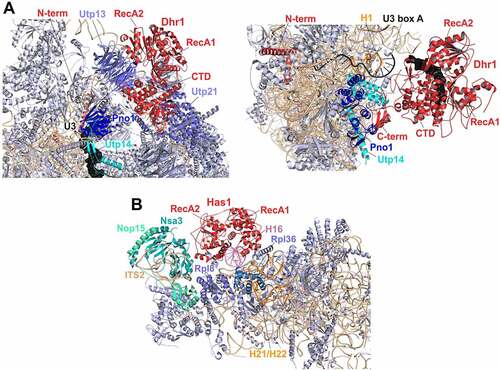
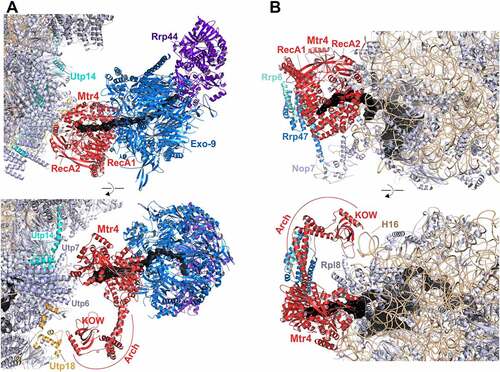
Figure 5. Cryo-EM structures of Mtr4 on 90S and pre-60S intermediates channelling its substrate RNAs for exosomal degradation.