Figures & data
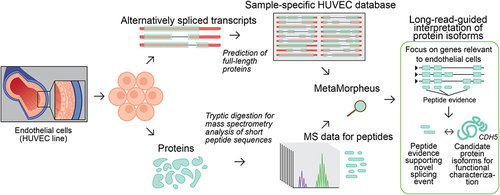
Figure 1. Characterization of isoform diversity in HUVECs through integration of long-read RNA-seq with mass spectrometry data (‘long read proteogenomics’). Transcripts are converted into a protein isoform database based on predicted open reading frames (ORFs) and the resulting database is searched against a sample-matched bottom-up mass spectrometry (MS) dataset. The peptide identifications can be used to support the expression of isoform candidates related to endothelial pathways.
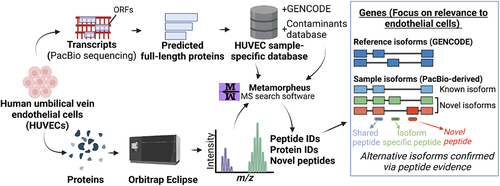
Figure 2. Characterization of transcript isoform diversity in HUVECs via long-read RNA-Seq. (A) Schematic of the long-read RNA-seq analysis pipeline. (B) Transcripts and genes identified from PacBio long-read RNA-seq. The number of known (blue) and novel isoforms (green and orange) are shown. (C) Transcript abundance distribution for known (FSM) versus novel transcripts (NIC, NNC), with dashed lines representing median abundance values in full-length read counts per million (CPM) for each category (FSM = 2.4, NIC = 1.5, NIC = 1.3). (D) Distribution of the number of genes expressing multiple isoforms. (E) Fraction of genes in which the most abundantly expressed isoform (“major isoform”) differs from the reference isoform (APPRIS principal isoform).
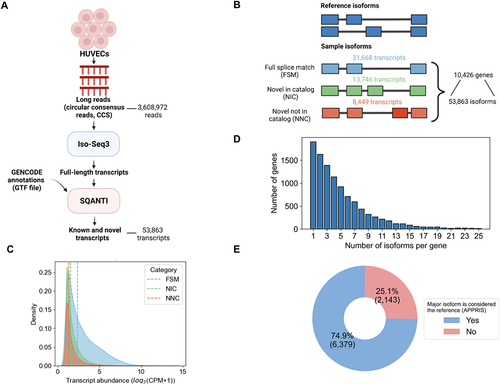
Table 1. Endothelial-relevant genes expressing multiple transcript isoforms in HUVECs.
Figure 3. Proteomic analysis of HUVECs using a customized long-read-derived protein isoform database. (A) Steps involved in the generation of a HUVEC sample-specific database. (B) Parallel long-read RNA-seq and MS proteomic data collection from HUVECs. (C) Correlation between estimated RNA and protein expression levels. PSM, peptide spectral match; CPM, full-length read counts per million. (D) Comparison of proteomic search results between the reference and HUVEC sample-specific database.
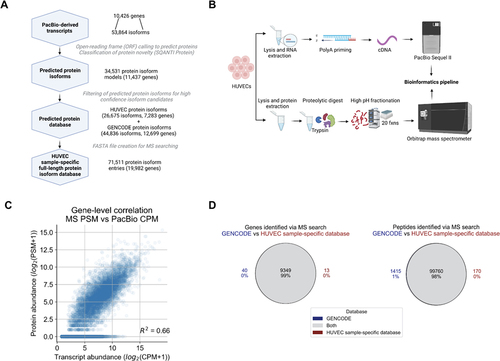
Table 2. Composition of the HUVEC sample-specific database.
Figure 4. Protein isoforms analysed based on peptides identified via mass-spectrometry (MS). (A) Scenarios of differing protein isoform detection precision when evidenced by peptides identified from MS. Only genes with multiple protein isoforms in the database are included, and 1,904 genes that express only one isoform were excluded. (B) A protein isoform confirmed with a uniquely mapping peptide LNE, for SRSF5, a splice factor that regulates transcripts of VEGF-A. (C) Two protein isoforms of TPM2 are confirmed with uniquely mapping peptides TID, AIS, and YKA. In B and C PacBio-derived protein isoform label follows this format: <Gene>|<PB accession>|<SQANTI Protein class>|<CPM>.
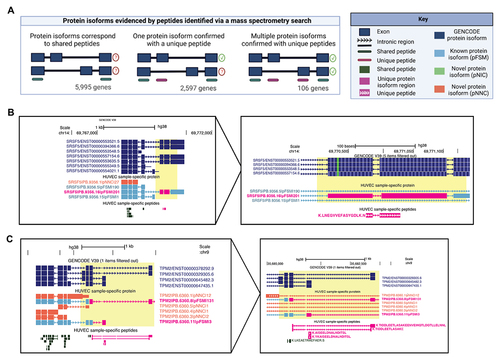
Figure 5. Nomination of protein isoforms when incorporating long-read data. (A) Scenarios of protein isoform candidates nominated for expression when transcript abundance from the long-read RNA-seq information is incorporated. (B) CDH5 gene, involved in endothelial pathways demonstrating a scenario of ambiguous protein isoforms identified only by shared peptides, but incorporation of long-read RNA-seq data suggests the expression of three moderately expressed protein isoforms (PB.10443.1, PB.10443.9 and PB.10443.71). (C) PECAM1 gene, involved in endothelial pathways demonstrating an example where one protein isoform is identified via a unique peptide (PB.1123.25), SDS, while the remaining protein isoforms are supported by shared peptides. Abundance information from long-read RNA-seq suggest expression of (PB.11293.1 and PB.11293.7). In B and C, PacBio-derived protein isoform label follows this format: <Gene>|<PB accession>|<SQANTI Protein class>|<CPM>. For B and C, low abundance protein isoforms (<25 CPM) are not shown.
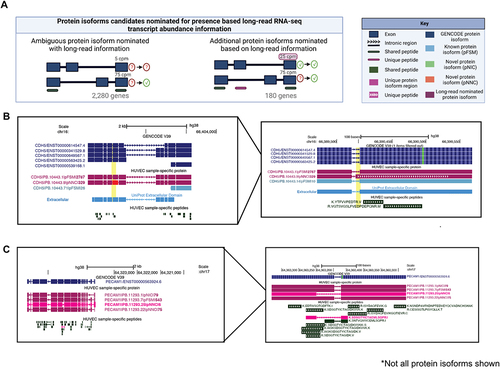
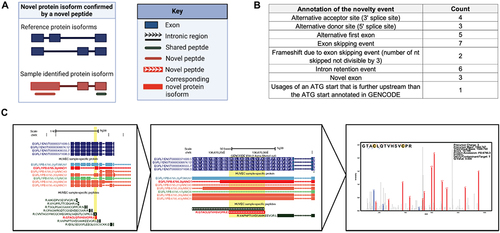
Figure 6. Novel protein isoforms discovered via unique peptides. (A) Novel protein isoform confirmed by identified novel peptides. (B) Table of the frequency of events supported confirmation of a novel peptide. (C) Novel peptide found for a protein isoform of endothelial gene EGFL7. Novel peptide and corresponding protein isoform shown in red, which supports a frameshift event for the protein isoform PB.6795.3.