Figures & data
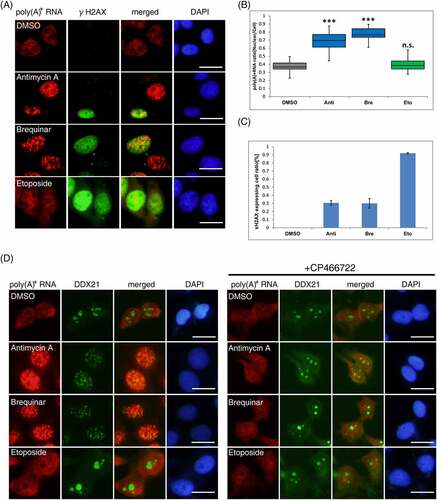
Figure 1. Screening of inhibitors for perturbing nuclear poly(A)+ RNA metabolism.
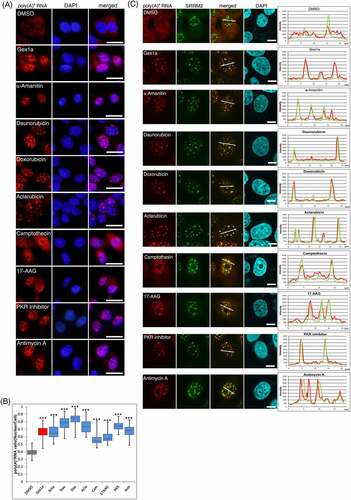
Figure 2. De novo pyrimidine synthesis pathway perturbs poly(A)+ RNA metabolism.
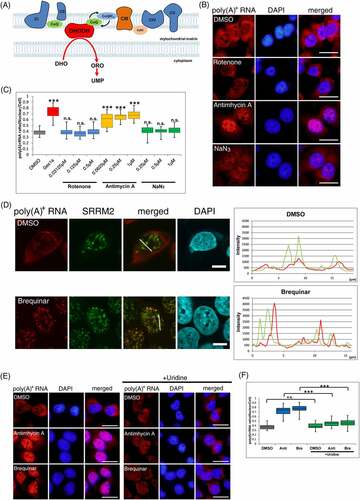
Figure 3. ATM activation is required to elicit nuclear accumulation of poly(A)+ RNA by inhibitors of the respiratory chain complex III and DHODH.
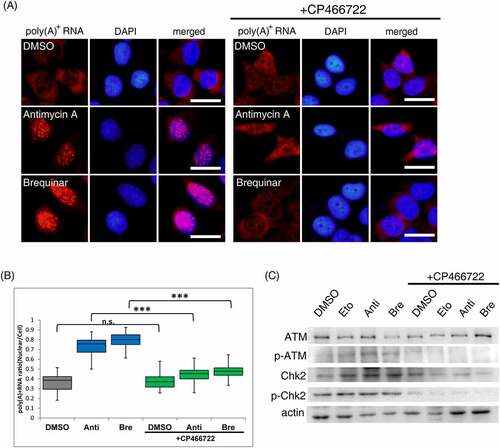
Figure 4. Relocalization of the nucleolar component by DHODH inhibition.