Figures & data
Table 1. CCR5 sgRNA position on chromosome 3 and specificity score.
Figure 1. Target sequences for gRNAs within the human CCR5 gene. Schematic of the locations of the target sequences for gRNAs (20 base protospacer sequence; yellow highlighting) and PAM sequences (light blue highlighting) relative to the location of the region that would be deleted in the 32 mutation (purple highlighting), within a 1,000 base pair segment of exon 2 of the human CCR5 gene located on chromosome 3.
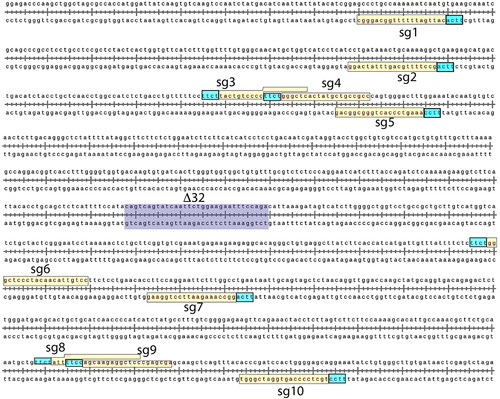
Figure 2. Cleavage activity of PlmCas12e is dependent on both gRNA length and target location.
Figure 3. Location of nine terminal PAM bases that were changed to assess CasX2 PAM preference. Four different CCR5 gene fragments of 1,114 bp (gBlocks) were synthesized to include nine of the ten gRNA target regions. The terminal PAM base for each gRNA was changed to either an A, C, G or T in each gBlock. Each of the four gene fragments were separately cloned into the pcDNA3.1 vector and then restriction enzyme digested with NheI and XhoI to yield a 1,074 bp target. In the CCR5 gene region of interest, four PAM regions were located on the (+) strand and five PAM regions were located on the (-) strand. For clarity, only the (+) strand (5’→ 3”) of the CCR5 sequence is illustrated here, where a yellow highlighted N indicates a PAM terminal base on the (+) strand, and a grey highlighted N indicates a PAM terminal base on the (-) strand. The blue highlighted region is the wild-type sequence of CCR5 that would be deleted in the CCR5-32 mutation. The sgRNAs 1, 2, 3 and 5 bind upstream of the CCR5-
32 region and sgRNAs 6, 7, 8, 9 and 10 bind downstream.

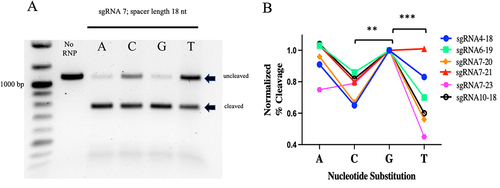
Figure 4. The terminal PAM base influences PlmCas12e cleavage activity.
Supplemental Material
Download Zip (2.3 MB)Data availability statement
The data that support the findings of this study are included in the manuscript.
