Figures & data
Figure 1. Chemical mechanisms of base excision from nucleic acids. A) Acid-induced hydrolysis relies on protons or hydrogen ions to protonate the adenine base which will trigger an electron delocalization around the dual ring eventually breaking the glycosidic bond. A nearby water molecule can resolve the oxocarbenium transition state. B) Methylation-induced base hydrolysis can occur when a base is methylated (for example by dimethyl sulfate, DMS) which will trigger an electron delocalization event severing the glycosidic bond. Like the acid-induced mechanism, a water molecule will resolve the transition state. C) in free radical base hydrolysis, the free electron displaces the hydrogen atom at the C1’ thereby producing superoxide. The oxocarbenium ion is resolved by a water molecule resulting in base excision and production of a carbonyl group on the C1’. D) an RNA N-glycosylase (pokeweed antiviral protein, PAP-I) removes the adenine by protonating the N−7 of the adenine with arginine 179. This will trigger an electron delocalization event thereby excising the glycosidic bond and producing an oxocarbenium ion that will be resolved by a water molecule directed by glutamic acid 176. Additional amino acids valine 73 and serine 121 help maintain the adenine in the correct orientation.
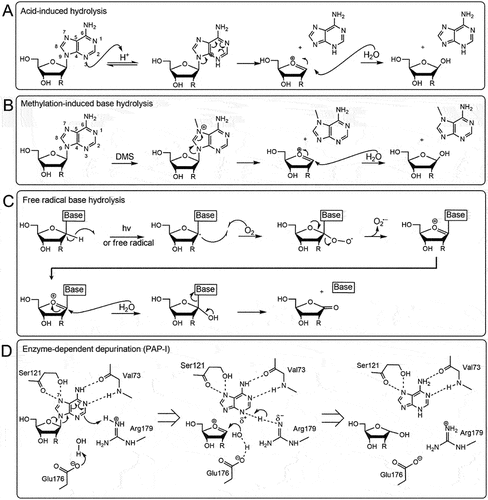
Figure 2. Reaction mechanism of aldehyde reactive probe (ARP). The abasic ribose exists in an unequal equilibrium between two conformations: the “closed” ring structure of the ribose and the “open” structure revealing an accessible aldehyde group. The aldehyde reactive probe is an amine, often biotinylated, that covalently binds with the exposed aldehyde group at the C1’ position of the abasic site.
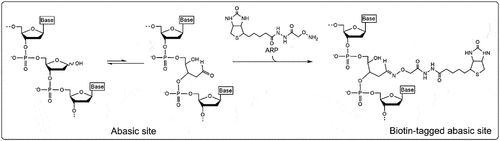
Figure 3. Abasic mRNA cross-linking with the ribosomal small subunit. A) an abasic nucleotide may cross-link with uS3, a small ribosomal subunit protein located near the mRNA entry site (PDB ID 4V6X). This cross-linking may stall the ribosome. mRNA is indicated as a red squiggly line and the site of cross-linking between Lysine 62 (shown in green) of uS3 and the abasic mRNA is circled. B) Detail of cross-linking between Lysine 62 of uS3 and the open conformation of an abasic ribose of the mRNA. Cross-linking involves a covalent Schiff base intermediate between the amino group of Lysine 62 and C1’ of the abasic site.
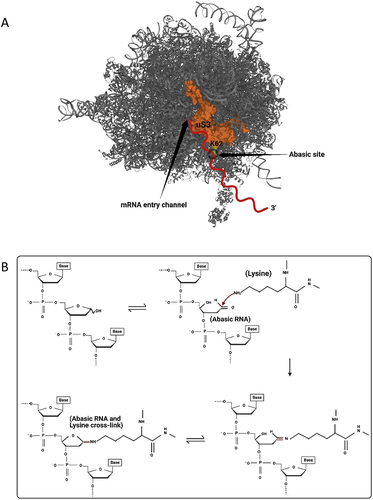
Figure 4. Abasic mRNA decay by the No-Go pathway. 1) Ribosome stalling with an abasic nucleotide at the A-site of the ribosome is recognized by Dom34p and Hbs1. 2) Hbs1 promotes binding of Dom34p to the A-site and 3) recruits ribosome-recycling factor ABCE1 for subsequent ribosome subunit dissociation. 4) Endonucleases, recruited through an unknown mechanism, cleave the mRNA 5) which is followed by exonuclease digestion from the 5’ and 3’ ends.
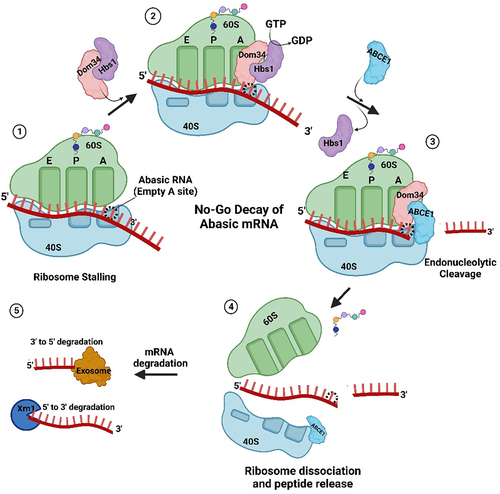
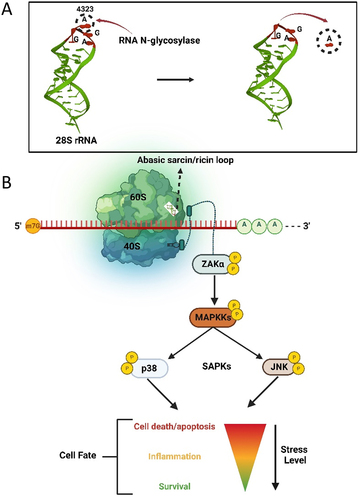
Figure 5. The ribotoxic stress response pathway. A) Hydrolysis of the adenine base (A4323) from the α-sarcin/ricin loop of 28S rRNA by an RNA N-glycosylase. B) Loss of a base from the α-sarcin/ricin loop (SRL) of the 28S rRNA is detected by the kinase ZAKα, which is bound to the ribosome. Through phosphorylation, damage to rRNA is signalled to downstream stress activated kinases JNK and p38, resulting in a proinflammatory response that may allow cell survival or terminate in apoptosis.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
