Figures & data
Figure 1. Summary of the canonical miRNA biogenesis pathway.
Biogenesis of miRNAs consists of the stepwise processing of pri-miRNAs in the nucleus and the cytoplasm. Briefly, pri-miRNAs fold into distinct hairpin-like structures that are recognized and cleaved by the Microprocessor, a heterodimer complex formed by DROSHA and DGCR8. The Microprocessor cleavage site (blue arrows) is defined by multiple structural and sequence motifs. The resulting product is exported into the cytoplasm via XPO5, for a second cleavage event by DICER (red arrows). The resulting duplex is loaded into one of the AGO paralogs for strand selection and assembly of mature RISC complexes with TNRC6. Recognition chambers inside AGO facilitate the recognition of target RNAs, and the subset of nucleotides involved in these facilitated pairings is known as the seed (cerulean circles) and the supplemental pairing regions (tangerine circles). Upon stable miRNA:target recognition, TNRC6 will mediate the recruitment of deadenylation complexes, leading to translational repression and decay of target mRNAs.
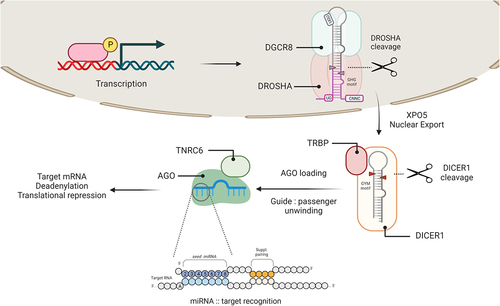
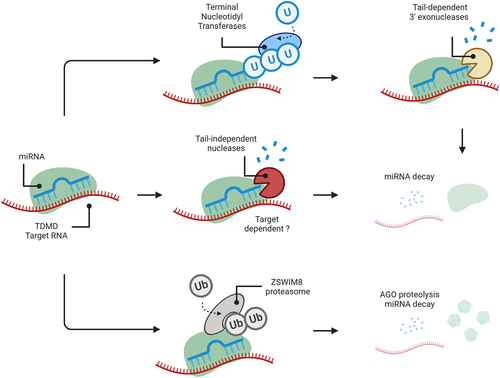
Figure 2. Summary of the mature miRNA decay pathways.
Decay of mature miRNAs can happen through multiple pathways. Up to date, three distinct mechanisms have been identified. (1) Tailing of miRNAs by terminal nucleotidyl transferases (TENTs) primes the decay via RNA nucleases. The example shows the decay via TUT4/7 mediated uridylation and DIS3L2. Similarly, TENT2 and TENT4B can mediate the adenylation that facilitates PARN mediated decay. (2) Other RNA nucleases, such as Tudor-SN, can also decay miRNAs independently of tailing. It is unclear whether certain target RNAs may facilitate the exposure of cleavable nucleotides. (3) ZSWIM8 can mediate the ubiquitination and proteolysis of AGO complexes. As a consequence, unprotected miRNAs can be degraded by cytoplasmic nucleases. In general, binding to target RNAs with extensive complementarity has been identified as a cue for most decay pathways.
Data availability statement
No data availability statement.
