Figures & data
Figure 1. Transient increase in expression of RNautophagy/DNautophagy-related genes by poly(I:C). A, B. relative levels of SIDT2 (A) or LAMP2C (B) mRNA in cells transfected with either poly(I:C) or mouse total RNA. C, D. relative levels of SIDT2 (C) or LAMP2C (D) mRNA in cells with poly(I:C) added directly into culture media. E, F. temporal change in expression levels of SIDT2 (E) or LAMP2C (F) mRNA in cells transfected with poly(I:C). (n = 3). ** p < 0.01, N.S., not significant.
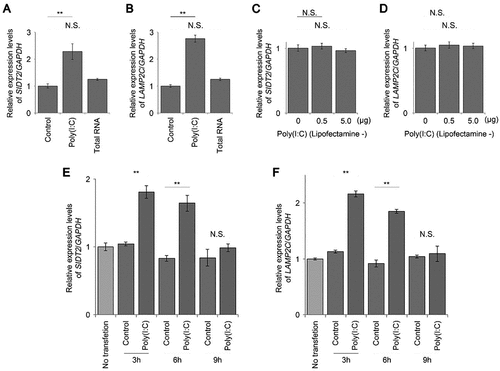
Figure 2. Poly(i:c) selectively upregulates the expression of RNautophagy/DNautophagy-related genes. A, B. dose-dependency in the expression levels of SIDT2 (A) or LAMP2C (B) mRNA in cells transfected with poly(I:C). C–F. relative levels of LAMP2A (C), LAMP2B (D), all LAMP2 isoforms (E), or LAMP1 (F) mRNA in cells transfected with poly(I:C). G. immunoblotting of SIDT2, LAMP2C, all LAMP2 isoforms, and LAMP1 protein in cells transfected with poly(I:C). H–K. quantification of respective gene products shown in G. (n = 3). * p < 0.05, ** p < 0.01, N.S., not significant.
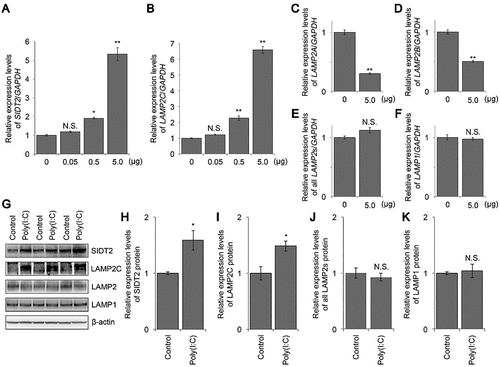
Figure 3. MDA5 mediates the upregulation of RNautophagy/DNautophagy-related genes expression by poly(I:C). A, B. relative levels of SIDT2 (A) or LAMP2C (B) mRNA in cells transfected with respective siRNA, followed by mock- or poly(I:C)-transfection. C, D. relative levels of SIDT2 (C) or LAMP2C (D) mRNA in cells transfected with respective expression vectors, followed by mock- or poly(I:C)-transfection. (n = 3). ** p < 0.01, N.S., not significant.
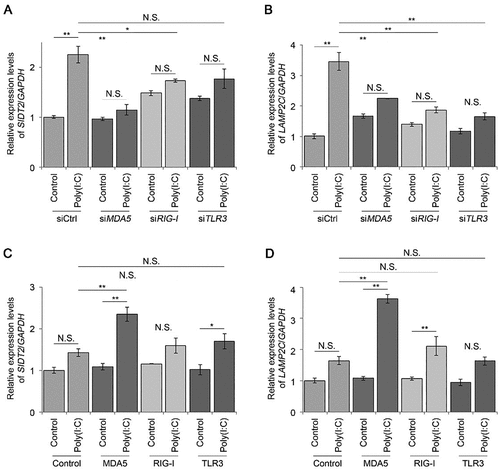
Figure 4. Viral RNA can be targeted for degradation via RNautophagy. A, B. relative levels of extracellular (A) and intracellular (B) viral RNA produced in WT or Sidt2 KO cells infected with JEV at MOI = 1. C. immunoblotting of extracellular viral protein produced by WT or Sidt2 KO cells infected with JEV at MOI = 1. D. a representative image of plaque assays using cultured media derived from WT or Sidt2 KO cells infected with JEV at MOI = 1. Plaques are surrounded by pen strokes. E. virus titre of cultured media derived from WT or Sidt2 KO cells infected with JEV at MOI = 1. F. relative levels of viral RNA in extralysosomal solution or lysosomal pellet following the incubation with isolated lysosomes in the absence or presence of ATP. G. relative levels of total viral RNA following the incubation with isolated lysosomes in the absence or presence of ATP. (n = 3). ** p < 0.01.
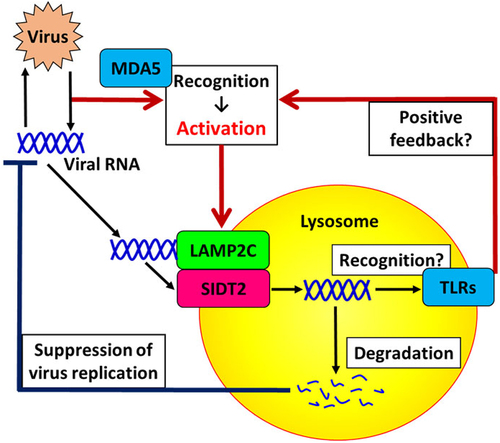
Figure 5. A schematic of our current model. Pattern recognition receptors (PRRs), such as MDA5, recognize pathogen-associated molecular patterns (PAMPs) such as dsRnas and upregulate RNautophagy/DNautophagy-related genes, to activate RNautophagy/DNautophagy. RNautophagy/DNautophagy contributes to clearance of viral nucleic acids, thereby suppressing virus replication. RNautophagy/DNautophagy may also contribute to the detection of PAMPs via endolysosomal TLRs by taking up viral nucleic acids into their recognition sites, which leads to induction of various anti-viral responses that may also include the activation of RNautophagy/DNautophagy as a positive feedback loop.
Supplementary Figures Yuuki Fujiwara 2023.docx
Download MS Word (466.3 KB)Data availability statement
The data generated or analysed in this study are available from the corresponding authors on reasonable request.
