Figures & data
Figure 1. Two mRNA identity features promote the nuclear export of most mRnas. A) Schematic of the splicing-dependent and GC-dependent mRNA export pathways. B) Illustration of how each feature is recognized by trans-factors, which promote mRNA export. Note that splicing, and likely GC-rich regions, recruit the EJC to both types of mRNA (see text for details).
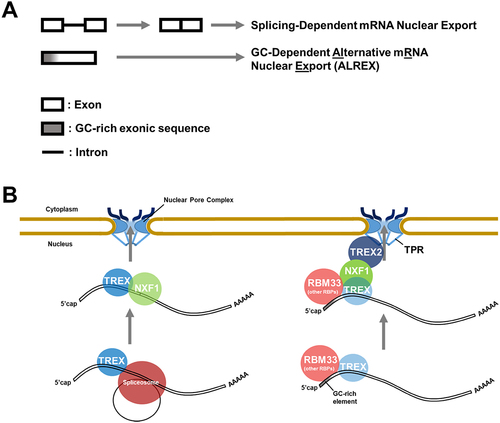
Table 1. Cis-elements and features that regulate mRNA quality control.
Figure 2. Sequence features of mRNAs. A-B) GC-content averaged over each exon (yellow) and intron (white) of all human protein-coding genes with 5 exons plotted from 5’ to 3’ ends. Note that in (A) each exon and intron metaplot was normalized, while in (B) they were adjusted to reflect the average length of each exon and intron of all genes in the dataset. C) Similar to (A) except that the average nucleotide-content of the coding strand was plotted from 5’ to 3’ ends.
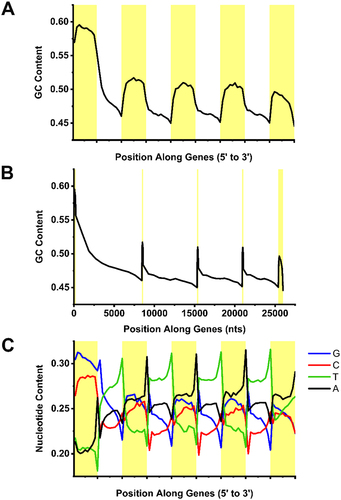
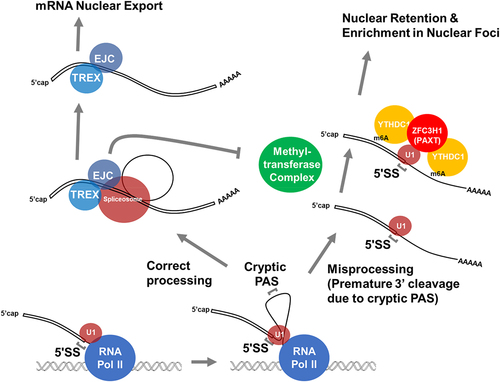
Figure 3. Misprocessing results in the preservation of splicing signals, which promote nuclear retention. Properly processed mRnas are compared to misprocessed mRnas that generate IPA transcripts. Note that the IPA transcript contains both an intact 5’SS due to the failure of splicing, and m6A modifications, due to the lack of deposited EJCs, which normally inhibits m6A modifications around the splice site. This could be due to the EJC sterically preventing the methylatransferase from accessing the mRNA (as depicted in the figure) or by the recruitment of demethylases such as ALKBH5.