Figures & data
Figure 1. Targeted mutagenesis of 25S rDNA led to mosaic mutation patterns on chromosome 4. (A) Schematical distribution of the rDNA copies on five Arabidopsis chromosomes and their associated VAR polymorphic rRNAs. (B) 3’-ETS polymorphism in the A. thaliana col-0 accession and the oligonucleotide’s binding positions for the detection and identification of mutated VARs in the genome. (C) Location of the targeted bases, consisting of the P-loop and helix-82 regions, in the secondary structure of the 25S LSU ribosomal RNA [Citation9] is indicated in magenta. Numbers 2600 and 2800 in cyan denotes the positions of nucleotides in 25S rRNA (D) Heteroduplex PCR products of three genotypes electrophoresed under 15% native PAGE conditions. (E) Quantification of the mutated and intact rDNA copies of the PCR products analysed in panel C (n = 6 linearly increasing PCR cycles). Data are shown as means ± SD among the total % of rDNA. The P-value indicates the statistical significance with the unpaired student t-test. (F) Cloning and sequencing of the heteroduplex PCR products of panel C resulting in mosaic mutation patterns in the rDNA. (G) Electrophoresis of the PCR products on 2.5% TTE gels from the three genotypes; PCR was carried out using the mut-F and ETS-R oligos, followed by cloning and sequencing of 33 independent clones – their linked VAR subtypes are summarized.
![Figure 1. Targeted mutagenesis of 25S rDNA led to mosaic mutation patterns on chromosome 4. (A) Schematical distribution of the rDNA copies on five Arabidopsis chromosomes and their associated VAR polymorphic rRNAs. (B) 3’-ETS polymorphism in the A. thaliana col-0 accession and the oligonucleotide’s binding positions for the detection and identification of mutated VARs in the genome. (C) Location of the targeted bases, consisting of the P-loop and helix-82 regions, in the secondary structure of the 25S LSU ribosomal RNA [Citation9] is indicated in magenta. Numbers 2600 and 2800 in cyan denotes the positions of nucleotides in 25S rRNA (D) Heteroduplex PCR products of three genotypes electrophoresed under 15% native PAGE conditions. (E) Quantification of the mutated and intact rDNA copies of the PCR products analysed in panel C (n = 6 linearly increasing PCR cycles). Data are shown as means ± SD among the total % of rDNA. The P-value indicates the statistical significance with the unpaired student t-test. (F) Cloning and sequencing of the heteroduplex PCR products of panel C resulting in mosaic mutation patterns in the rDNA. (G) Electrophoresis of the PCR products on 2.5% TTE gels from the three genotypes; PCR was carried out using the mut-F and ETS-R oligos, followed by cloning and sequencing of 33 independent clones – their linked VAR subtypes are summarized.](/cms/asset/f9ba255d-616a-4a00-a5d2-e626980f82f7/krnb_a_2298532_f0001_oc.jpg)
Figure 2. Mutation caused dosage-dependent growth defects linked to a single locus. (A) Seedling growth phenotypes of segregating heterozygous plants. Scale bar: 10 mm (B) root length variations analysed during early growth (n = 18, BG; n = 29, ploop KD ±; n = 16, ploop KD -/- vertically grown seedlings). (C) Rosette morphology of soil-grown mutants. Scale bar: 30 mm (D) rosette diameters of mutants (n = 18 replicates for each genotype). (E) Seed morphology of ploop KD -/- mutants. (F) Genotype-phenotype correlation of 120 randomised seedlings based on the phenotypical segregation as in figure 2A and further genomic PCR analysis of the ploop allelic band (+, present; -, absent) of all seedlings. (G) Venn analysis of the identified proteome based on 4 biological replicates of 10 DAS BG and ploop KD -/- seedlings. (H) Bicluster analysis of the abundance of varying levels of GO terms-based classified proteins between 4 replicates. Data in (B) and (D) are presented as means ± SD.
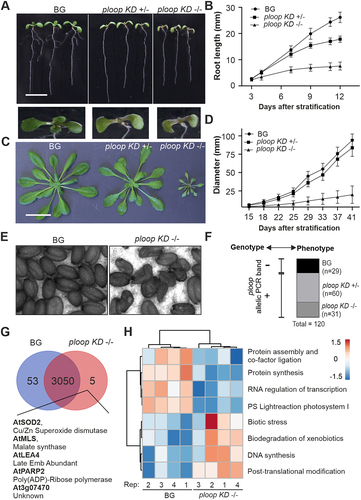
Table 1. List of proteins distinctly absent in ploop KD -/- seedlings.
Figure 3. Mutation caused dosage compensation from the usually inactive NOR2 locus leading to other rRNA defects.
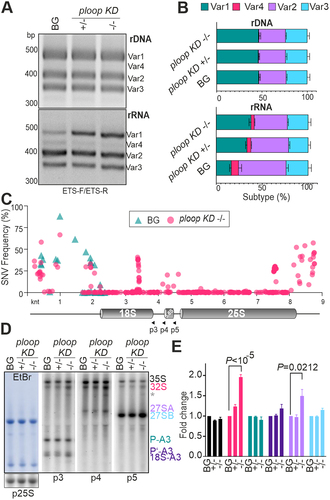
Figure 4. Mutated copies are not incorporated into the translating polysomes. (A) Absorbance profiles at 254 nm of the floral tissues from BG (black) and ploop KD ± (magenta) seedlings plotted with time of detection from top to bottom following sucrose-density gradient centrifugation. (B) The RNA from the corresponding fractions in panel a for both genotypes was purified, reverse transcribed with the 25S rRNA-specific oligo and the PCR products with the mutation-specific forward oligo and 25S-specific reverse oligo resolved by 12% native PAGE gels. The black arrow denotes the PCR-run through product and the magenta arrow indicates the mutated rRNA product. (C) The proteins from the fractions in panel a of both genotypes were resolved on 10% SDS-PAGE and blotted with the indicated antibodies shown on the right-hand side.
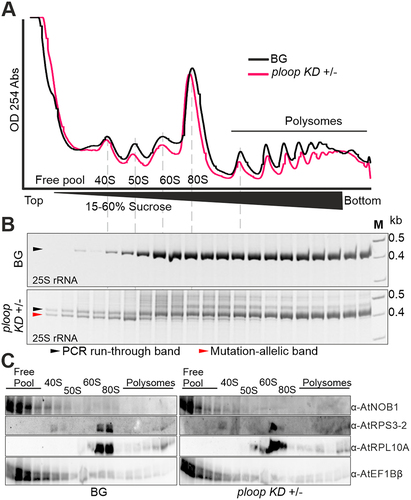
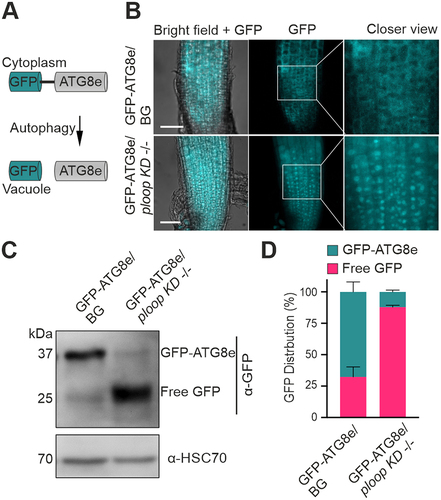
Figure 5. Mutants display elevated autophagic flux. (A) Schematics of the GFP-ATG8e fusion protein localisation under routine growth and autophagic conditions. (B) Localisation of the GFP signals from the GFP-ATG8e transgenic fusion proteins in the BG (top) and ploop KD -/- (bottom) seedlings. Scale bars: 50 µm (C) immunoblotting analysis with the anti-GFP antibodies of the GFP-ATG8e levels in the BG and ploop KD -/- seedlings. Anti-HSC70 blotting served as the loading control. (D) Quantification of the full length GFP-ATG8e and free GFP signals from panel C represented in relation to the total levels after normalisation to the HSC70 loading control (n = 3 biological replicates). Data are presented as means ± SD % of each GFP variant to the total GFP signal intensity after normalizing with the HSC70 loading control signals.
Supplemental Material
Download MS Word (5.1 MB)Dataset S1.xlsx
Download MS Excel (5.3 MB)Dataset S2.xlsx
Download MS Excel (20.1 KB)Data availability statement
The RNA-seq data presented in this study are deposited in GEO repository under accession no. GSE213764 and the proteome data are deposited in PRIDE repository with accession no. PXD035623.
