Figures & data
Figure 1. Haematopoiesis and the development of haematological malignancies. A simplified schematic representation of haematopoiesis demonstrating the origin of myeloid and lymphoid lineages. Cancer types described in this review (marked in blue), develop from the cells at a particular step of haematopoiesis that is in the bone marrow (highlighted in purple) or in the lymphatic system (in green). Arrows indicate the direction of cell differentiation. See text for abbreviations.
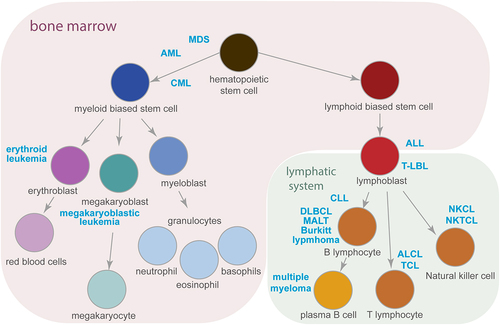
Figure 2. The ARE-BP axis is a functionally related network crucial for leukaemia development and chemotherapy resistance. Evidence-based interaction network retrieved from the string-db.org database. A full STRING network is presented; edges set to confidence (line thickness indicates the strength of data support); sources of interaction: experiments, co-occurrence, and co-expression; minimum required interaction score confidence set to low (0.15). GeneOntology enrichment analysis shows proteins involved in the regulation of mRNA stability (GOBP:0043488 in red), regulation of translation (GOBP:0006417 in blue) and localized in cytoplasmic ribonucleoprotein (RNP) granules (GOCC:0036464 in green).
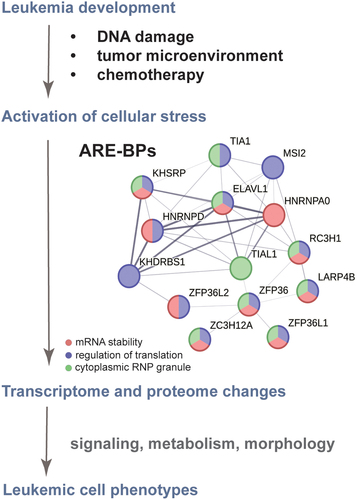
Figure 3. ARE-BPs recognize sequences in RNA called AU-rich elements (AREs). A) Sequence motif logos representing the most frequent nucleotide in each position recognized by the indicated ARE-BP from the ENCORE/ENCODE project database for cell-free pulldown experiments (RNA bind-n-seq). B) Results from crystal structure analysis of RNA binding domains (in green/blue) of selected proteins in complex with ARE (in brown). From the left are the two N-terminal RRM domains of HuR bound to poly(U) RNA from crystal structure PDB ID: 4ED5; the ROQ domain from murine roquin-1 bound to a 23-mer tnf-CDE RNA from X-ray structure PDB ID:4QI2; the KH domains of T-STAR dimer bound to AAAUAA RNA from structure PDB ID: 5ELS.
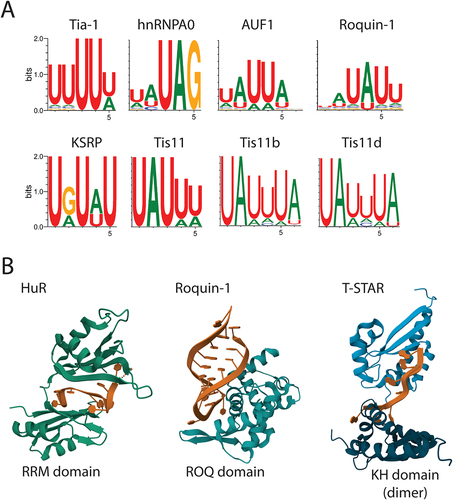
Table 1. Summary of information about the ARE-binding proteins discussed in this review. Provided references cite studies of the ARE-BP in the context of leukaemia or lymphoma. See text for abbreviations. *direct link to cancer development has not been shown.
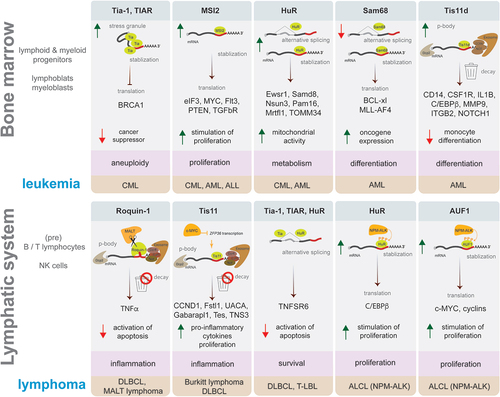
Figure 4. Examples of ARE-binding proteins role in leukaemia and lymphoma development. Schematic representation in each box presents an example of ARE-BP activity discussed in this review that affects a process supporting malignancy (highlighted in pink), which plays a role in the development of leukaemia (upper panel) or lymphoma (lower panel) subtype (highlighted in brown). ARE-BPs change the level of proteins causing stimulation (green arrow up) or inhibition (red arrow down) of cellular processes, by modulating RNA processing like alternative splicing, stabilization, decay, translation, and formation of stress granules or p-bodies. Key to symbols: mRNA decay (trash can), ARE-BP (green), oncogene (dark yellow), CCR4-NOT deadenylase complex with exosome (brown), Dcp2 decapping enzyme (grey), AU-rich element (red line), mRNA 5’ cap (grey dot).