Figures & data
Figure 1. Analytical pipeline used to characterize circRNAs.
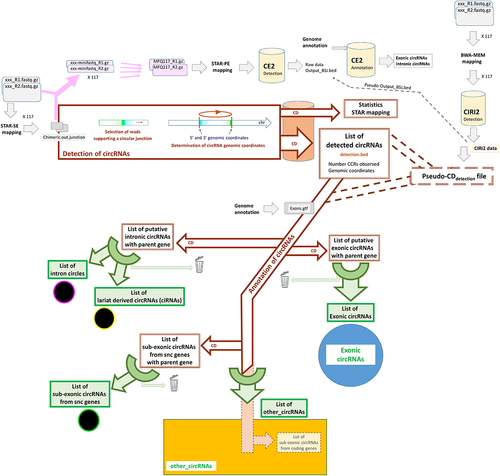
Figure 2. Overview of circRNAs detected in the 117 samples considered.
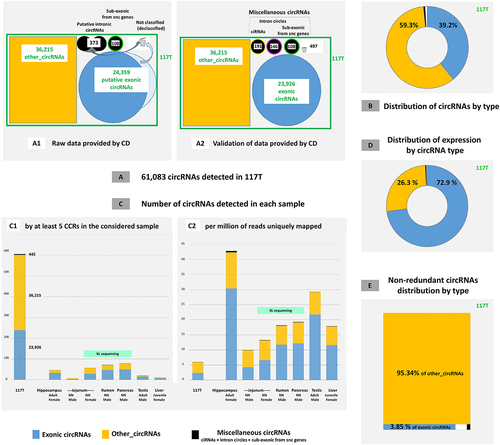
Figure 3. Analysis of circRNAs detected in mRNAseq.
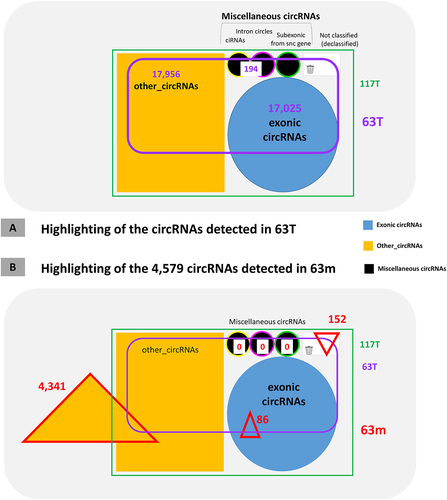
Figure 4. Analysis of circRNAs detected by CIRI2.
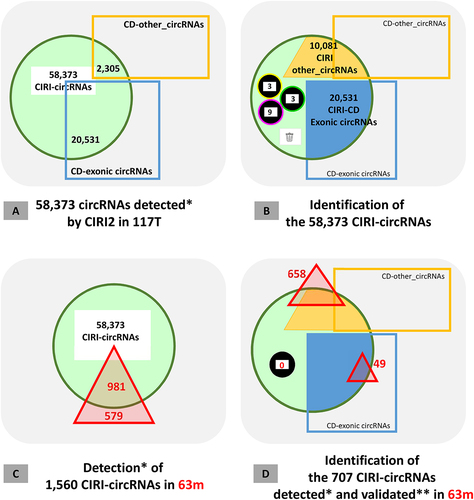
Figure 5. Analyses of the possible presence of 23,737 exonic circRNAs in bovine tissues/samples.
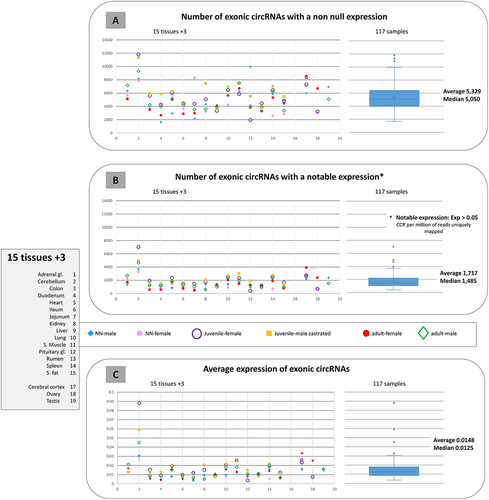
Figure 6. Expression analysis of 23,737 exonic circRNAs in four samples.
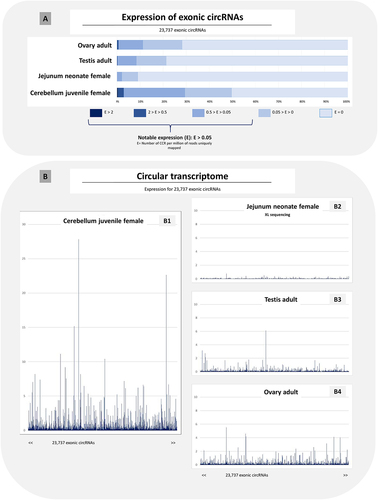
Figure 7. Benchmarking of 23,737 reliable exonic circRNAs and additional exonic circRNAs.
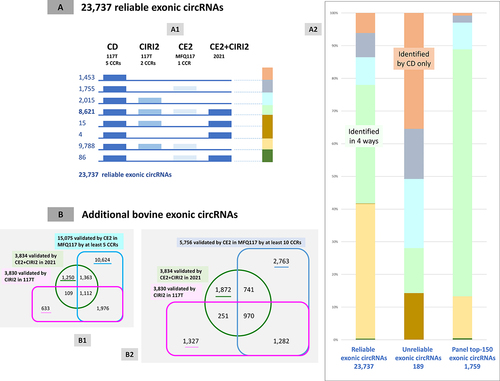
Figure 8. Hierarchical clustering analysis (HCA).
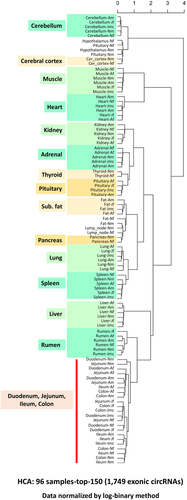
Figure 9. Principal component analyses (PCA).
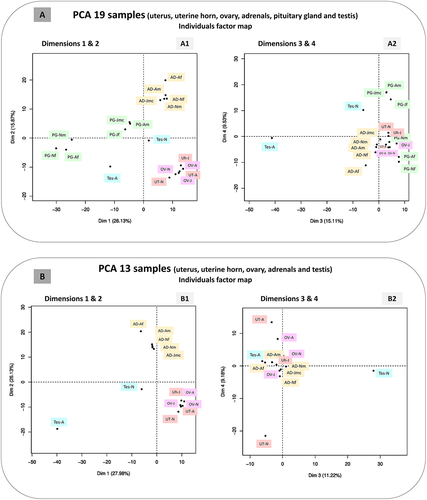
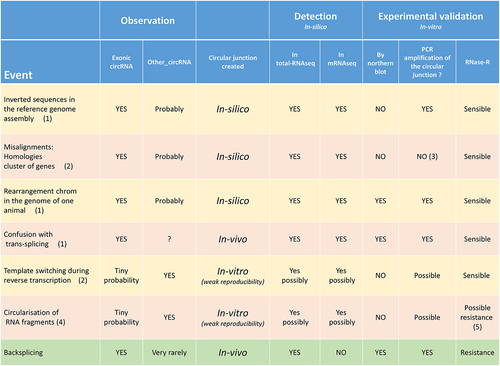
Figure 10. List and characteristics of different events leading to the formation of a circular junction.
Atab.pdf
Download PDF (1.9 MB)Res_Adoc.pdf
Download PDF (2.8 MB)MM_Adoc.pdf
Download PDF (2.4 MB)Data availability statement
All data obtained concerning exonic and intronic circRNAs are available in several tables (all Ext_Atab) deposited in Dataverse repository (doi: 10.57745/XORQHK). The list of other_circRNAs is not available, as we were unable to distinguish between reliable and unreliable other_circRNAs. List of exons generated by the BovReg consortium and used in this study is available on upon request from CC. Datasets generated by the BovReg consortium and analysed during the current study are listed in Atab-1. We built 117 paired mini-fastq files (R1 and R2) to provide all sequences allowing a global and rapid characterization of circular RNAs present in bovine tissues (doi: 10.57745/IUJ40P).
