Figures & data
TABLE 1 Superarray Macroarray Table for 96 human stress and toxicity genes tetra-spotted on the arrays
TABLE 2 Indirubin-3′-treatment regimens
TABLE 3 List of PCR primers, cycling conditions and amplicon size for each gene used to validate the macroarray results from treating U937 cells with indirubin-3′-monoxime
FIG. 1 CYP1A1 protein expression in PMA-differentiated U937 cells following exposure to indirubin-3′-monxime treatment for 48 hrs. Differentiated U937 cells were treated with indirubin-3′-monoxime, lysed, and subjected to SDS-PAGE/immunoblots steps and probed for CYP1A1 protein (a) or β-actin protein (b). Lane 1, carrier control; lane 2, 10 μ M; lane 3, 1 μ M; lane 4, 0.1 μ M; lane 5, 0.01 μ M.
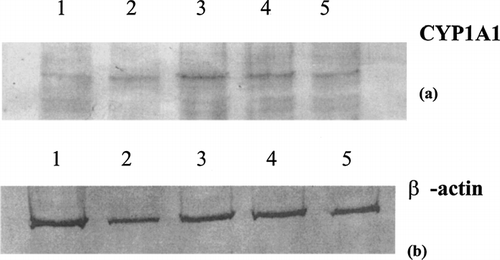
FIG. 2 Cytotoxicity of 10 μ M and 1 μ M indirubin-3′-monoxime to U937 cells under different stages of differentiation and activation. Regimes, A representing monocytes, B representing macrophages, C representing differentiation process, and D representing activated macrophages. Data represent means of three experiments +/− standard error. Viability as determined by MTT assay. Tributyltin chloride was included as a positive control showing cellular toxicity.
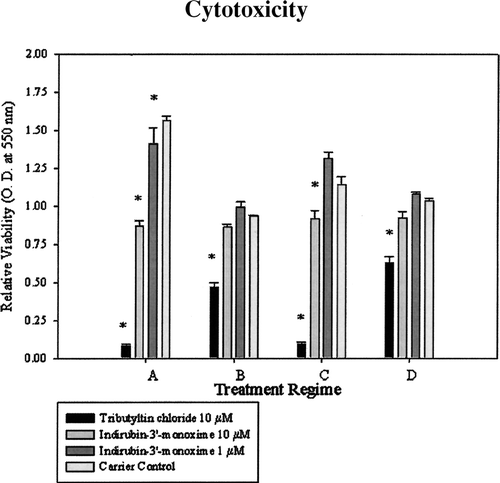
TABLE 4 Altered gene expression in U937 cells following exposure to 1 μ M indirubin-3′-monoxime (regimen A) as determined by macroarray analysis
TABLE 5 Altered gene expression in PMA-differentiated U937 cells (regimen B) following exposure to 1 μ M indirubin-3′monoxime as determined by macroarray analysis
TABLE 6 Altered gene expression in U937 cells exposed to 1 μM indirubin-3′-monoxime and then differentiated with PMA (regimen C) as determined by macroarray analysis
TABLE 7 Altered gene expression in PMA-differentiated and LPS-activated U937 exposed to 1 μ M indirubin-3-monoxime (regimen D) as determined by macroarray analysis
FIG. 3 Photograph of RT-PCR for COX-2, CYP1A1, MIP-1β, and GAPDH expression in U937 cells treated with 1 μ M indirubin-3′-monoxime under four different regimens. Regimen A, U937 cells treated with indirubin-3′-monoxime (IR) or carrier control (CC); Regimen B, effects of indirubin-3′-monoxime (IR) and carrier control (CC) on PMA-differentiated U937 cells; Regimen C, effects of indirubin-3′-monoxime (IR) and carrier control (CC) on PMA-induced differentiation of U937 cells; and Regimen D, effects of indirubin-3′-monoxime treatment (IR) and carrier control (CC) on differentiated and LPS-activated cells.
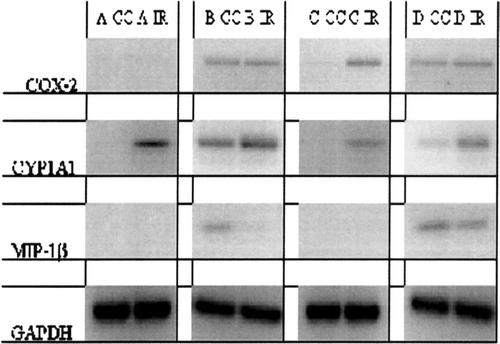
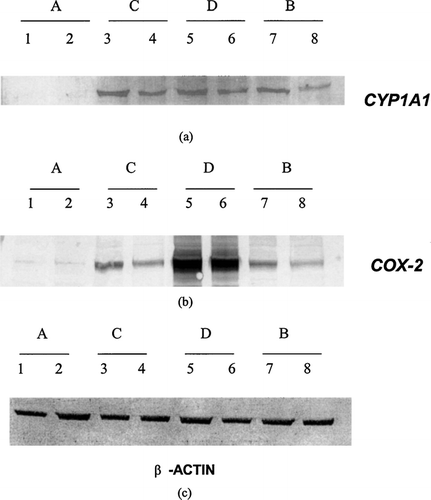
FIG. 4 Immunoblots for CYP1A1 (a), COX-2 (b), and β -actin (c) in U937 cells treated with 1 μ M indirubin-3′-monoxime under four different regimens. Regimen A, U937 cells treated with indirubin-3′-monoxime (lane 1) and carrier control (lane 2) for 24 hrs; Regimen C, effects of indirubin-3′-monoxime (lane 3) and carrier control (lane 4) on PMA-induced differentiation of U937 cells; Regimen D, effects of indirubin-3′-monoxime (lane 5) and carrier control (lane 6) on differentiated and LPS-activated cells; and Regimen B, effects of indirubin-3′-monoxime (lane 7) and carrier control (lane 8) on PMA-differentiated U937 cells.