Figures & data
TABLE 1 Primer sequences for qRT-PCR
FIG. 2 (A) Changes at 6 hr of genes differentially regulated at both 6 and 24 hr. The top bar for each gene is the data from the microarray, the middle bar is the data from the same mRNA analyzed by quantitative PCR, and the bottom bar represents the data from an independent experiment involving four treated and four control animals analyzed by quantitative PCR. By microarray, it was found that 42 genes were significantly differentially regulated at both 6 hr and 24 hr. In this figure, the focus is on 9 of these genes that are involved in cell stress and the promotion of liver regeneration or cell growth. (B) Changes at 24 hr of genes differentially regulated at both 6 and 24 hr. Graphed are the same 9 genes graphed in the same manner but at the 24 hr timepoint. Data for both graphs are expressed as log2 values, and all gene expression relative to GAPDH. All qRT-PCR were repeated 2 to 3 times, and run in triplicate. Standard error bars indicate variability between PCR runs.
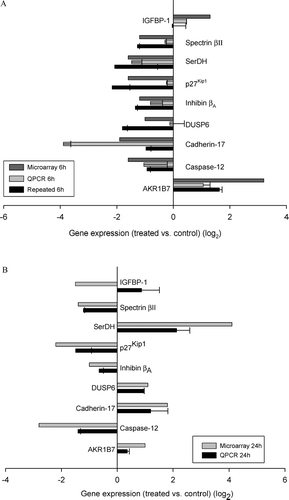
TABLE 2 Brief description of genes involved in liver regeneration, Cell-cycle promotion, intercellular adherence, and cell polarization
TABLE 3 Brief description of genes involved in oxidative stress, inflammation and Cytotoxicity, and Liver regeneration
FIG. 3 Genes differentially regulated at 6 hr only. Shown here are the data on 12 genes of > 300 genes that were significantly different by microarray from control at 6 hr. These genes were chosen because they are associated with oxidative stress, inflammation, cytotoxicity or are promoters of liver regeneration. Similar to , the top bar for each gene is the data from the microarray, the middle bar is the data from the same mRNA analyzed by quantitative PCR, and the bottom bar represents the data from an independent experiment involving 4 treated and 4 control animals analyzed by quantitative PCR. Data are expressed as log2 values, and all gene expression relative to GAPDH. All qRT-PCR were repeated 2 to 3 times, in triplicate or duplicate. Standard error bars indicate variability between PCR runs.
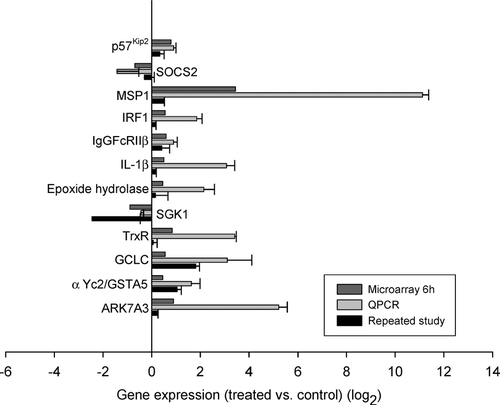
TABLE 4 Genes differentially regulated at 6 hr that were of interest to our study objective, but on which qRT-PCR was not performed
TABLE 5 Fold-changes in Hepatic gene expression induced by treatment with SMX, either 6 or 24 hr after SMX treatment