Figures & data
Figure 1. trnL intron and trnL-F regions (Amirahmadi et al., Citation2010)
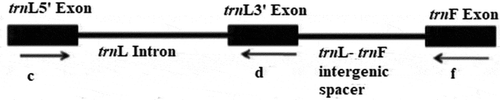
Table 1. Cycles and conditions of cpDNA trnL intron and trnL-F region PCR reactions
Table 2. Length and A + T and G + C contents of trnL intron sequences of Laurus nobilis populations
Table 3. Pairwise sequence distances among Laurus nobilis populations taxa for trnL intron data using MEGA 6.0 software distance matrix
Table 4. Tajima’s Neutrality Test Values based on trnL intron of eight date Laurus nobilis populations MEGA6
Table 5. Maximum likelihood estimate of substitution matrix
Table 6. Length and A + T and G + C contents of cpDNA trnL-F sequences of Laurus nobilis populations
Table 7. Pairwise sequence distances among some Laurus nobilis populations for cpDNA trnL-F data using MEGA 6.0 software distance matrix
Table 8. Tajima’s Neutrality Test Values based on trnL-F of eight date Laurus nobilis populations MEGA6
Table 9. Maximum likelihood estimate of substitution matrix
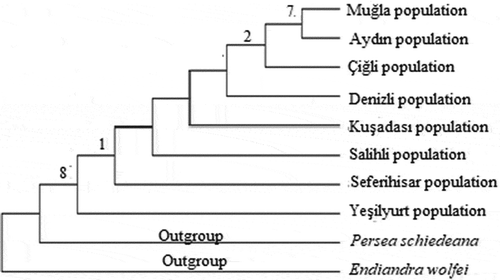
Figure 3. Phylogenetic tree of Laurus nobilis populations trnL intron sequences constructed using maximum likelihood method with MEGA 6.0
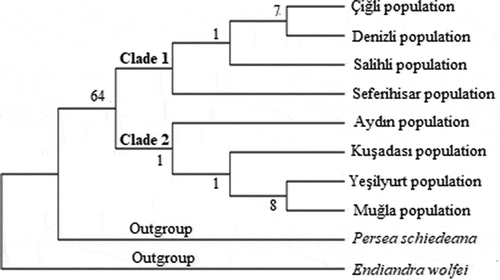
Figure 4. Phylogenetic tree of Laurus nobilis populations trnL intron sequences constructed using neighbor joining method with MEGA 6.0
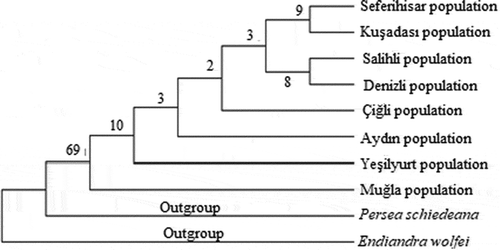
Figure 5. Phylogenetic tree of Laurus nobilis populations trnL-F sequences constructed using maximum likelihood method with MEGA 6.0
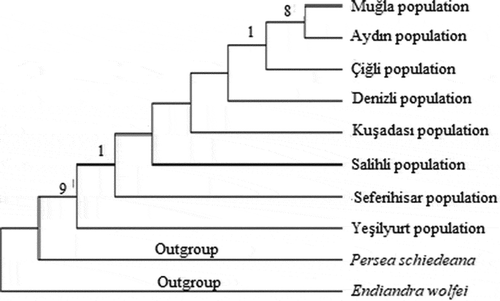
Figure 6. Phylogenetic tree of Laurus nobilis populations trnL-F sequences constructed using neighbor joining method with MEGA 6.0