Figures & data
Figure 1. PSMD10 enhances autophagy in starvation. (A, B) Evaluation for LC3B-II and SQSTM1 alteration indicative of autophagy induction in SMMC-7721-luc, SMMC-7721-PSMD10, or HCC-LM3 cells infected with adenovirus-mediated siRNA against GFP (Ad-siGFP) or PSMD10 (Ad-siPSMD10) treated with Earle's balanced salt solution (EBSS) for up to 24 h (A) or EBSS plus chloroquine (CQ) for 12 h (B). (C) SMMC-7721 cells were transiently cotransfected with pcDNA3.1 or PSMD10, or HCC-LM3 cells with siCon or siPSMD10. After 18 h transfection, cells were retransfected with pGFP-LC3B plasmid and subjected to EBSS treatment for the indicated time before observation for GFP-LC3B puncta under a fluorescence microscope. The percentage of cells with accumulation of GFP-LC3B in puncta was calculated in 3 random fields. (D) Representative images of LC3B staining in SMMC-7721-luc or SMMC-7721-PSMD10 cells infected with adenovirus-delivering mRFP-GFP-LC3B and treated with EBSS for 12 h. Quantification of autophagosome and autolysosome formation representing puncta staining sites per cell of 10 independent images. (E) Representative electron micrographs of autophagic vesicles or autophagosomes of (A) for up to 24 h. Arrows denote autophagosomes. Magnified image is shown. (F, G) Immunoblot for the indicated molecules in liver tissues isolated from 8-wk-old Psmd10-transgenic mice and littermates in a fed (F) or a fasted state for 24 h (G). (H) Immunoblot in liver tumors isolated from diethylnitrosamine (DEN) plus TCPOBOP-induced HCC in Psmd10-transgenic mice and littermates. Data represent the mean ±SD of 3 independent experiments (*P < 0.05, **P < 0.01).
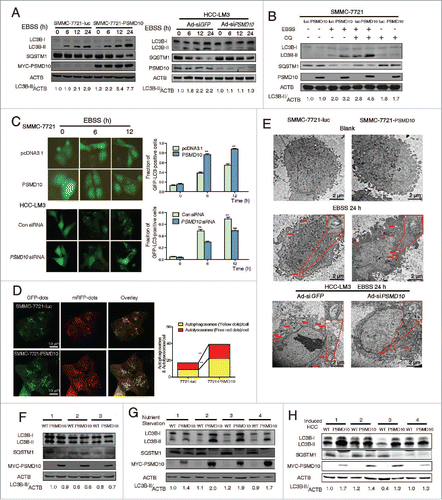
Figure 2. PSMD10 interacts with ATG7 to promote autophagy in EBSS starvation. (A) SMMC-7721-PSMD10 cells were EBSS-starved for the indicated time, and the cytosolic lysate was extracted for coimmunoprecipitation with anti-MYC, followed by probing with anti-ATG7. (B) The cell lysate from SMMC-7721 treated with EBSS for 12 h was extracted for coimmunoprecipitation with anti-PSMD10 or ATG7, followed by probing with anti-ATG7 or PSMD10. (C) HCC-LM3 cells treated with EBSS for 12 h were immunostained for endogenous PSMD10 and ATG7 to observe their colocalization, DNA was stained by 4',6-diamidino-2-phenylindole (DAPI). Representative confocal microphotographs are shown together with profiles of colocalization within the area of interest. (D) Different MYC-tagged PSMD10-expressing plasmids (mutants with each of the ankyrin repeats [dA1 to dA5], the N terminus [dN, deleted for amino acids 1 to 38], or the C terminus [dC, deleted for amino acids 204 to 226] deleted), were transfected into SMMC-7721 cells, and immunoprecipitated and immunoblotted with ATG7 or MYC-tag antibodies. (E) HEK-293 cells cotransfected with MYC-tagged PSMD10 and HA-tagged ATG7 (WT), ATG571S, ATG7CΔ, or a combination of the 2 ATG571S, CΔ, were immunoprecipitated and immunoblotted with the indicated antibodies. (F) LC3B-II accumulation was detected in SMMC-7721 cells transfected with MYC-tagged PSMD10 mutants (dA3 to 5, dC) or a PSMD10 complete plasmid after EBSS treatment for 12 h. (G) Coimmunoprecipitation with anti-HA was performed in SMMC-7721 cells transfected with HA-tagged ATG7, MYC-tagged PSMD10 mutants (dA3 to 5, dC), or a PSMD10 complete plasmid, and EBSS treated for 12 h, followed by probing for the indicated molecules.
![Figure 2. PSMD10 interacts with ATG7 to promote autophagy in EBSS starvation. (A) SMMC-7721-PSMD10 cells were EBSS-starved for the indicated time, and the cytosolic lysate was extracted for coimmunoprecipitation with anti-MYC, followed by probing with anti-ATG7. (B) The cell lysate from SMMC-7721 treated with EBSS for 12 h was extracted for coimmunoprecipitation with anti-PSMD10 or ATG7, followed by probing with anti-ATG7 or PSMD10. (C) HCC-LM3 cells treated with EBSS for 12 h were immunostained for endogenous PSMD10 and ATG7 to observe their colocalization, DNA was stained by 4',6-diamidino-2-phenylindole (DAPI). Representative confocal microphotographs are shown together with profiles of colocalization within the area of interest. (D) Different MYC-tagged PSMD10-expressing plasmids (mutants with each of the ankyrin repeats [dA1 to dA5], the N terminus [dN, deleted for amino acids 1 to 38], or the C terminus [dC, deleted for amino acids 204 to 226] deleted), were transfected into SMMC-7721 cells, and immunoprecipitated and immunoblotted with ATG7 or MYC-tag antibodies. (E) HEK-293 cells cotransfected with MYC-tagged PSMD10 and HA-tagged ATG7 (WT), ATG571S, ATG7CΔ, or a combination of the 2 ATG571S, CΔ, were immunoprecipitated and immunoblotted with the indicated antibodies. (F) LC3B-II accumulation was detected in SMMC-7721 cells transfected with MYC-tagged PSMD10 mutants (dA3 to 5, dC) or a PSMD10 complete plasmid after EBSS treatment for 12 h. (G) Coimmunoprecipitation with anti-HA was performed in SMMC-7721 cells transfected with HA-tagged ATG7, MYC-tagged PSMD10 mutants (dA3 to 5, dC), or a PSMD10 complete plasmid, and EBSS treated for 12 h, followed by probing for the indicated molecules.](/cms/asset/cd3ab45f-7ed1-4390-b34c-d9e4205e9c79/kaup_a_1034405_f0002_c.gif)
Figure 3. PSMD10 promotes ATG7 transcriptional activation. (A) Protein contents of ATG7, BECN1, ATG12–ATG5 conjugation, and PSMD10 were examined by immunoblot in SMMC-7721-PSMD10 versus SMMC-7721-luc cells, and HCC-LM3-Ad-siPSMD10 vs. Ad-siGFP cells after EBSS treatment for the indicated times. (B, C) The effect of PSMD10 on ATG7 expression was analyzed by immunoblot in vivo, including 24-h fasting (B) or DEN-TCPOBOP-induced HCC models (C). (D) Representative micrographs of autophagic vesicles or autophagosomes are shown in SMMC-7721 cells after transient cotransfection with pcDNA3.1 or MYC-PSMD10 and a nonspecific siRNA control or siATG7 RNA #1. (E) SMMC-7721-luc or SMMC-7721-PSMD10 cells were exposed to EBSS and cycloheximide (CHX, 1 μg/ml), actinomycin D (ActD, 5 μg/ml), or MG132 (5 μM) for 12 h. ATG7 was determined by immunoblot. (F) Quantitative PCR was performed to measure the transactivity of ATG7 in stable SMMC-7721-luc or SMMC-7721-PSMD10 cells exposed to EBSS for 24 h. Data represent the mean ± SD of 3 independent experiments (*P < 0.05).
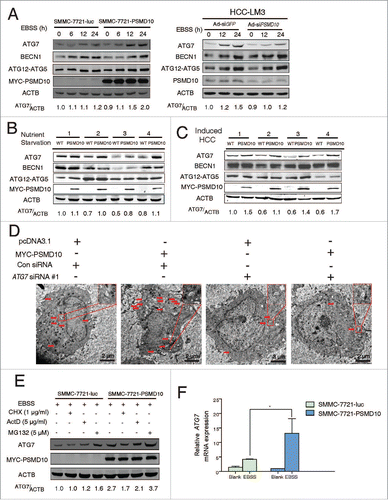
Figure 4. PSMD10 translocates into the nucleus and binds to the promoter of ATG7. (A) . PSMD10 translocates into the nucleus and binds to the promoter of ATG7. (A) Nuclear translocation of endogenous PSMD10 in HCC-LM3 was detected by immunostaining with anti-PSMD10 antibody after EBSS starvation for different time points as indicated. Arrows indicate nuclear localization of PSMD10. (B) Cell fractionation from HCC-LM3 cells was performed to analyze the cellular localization of endogenous PSMD10. Histone H3 and GAPDH were assessed as loading controls for nuclear and cytoplasmic proteins, respectively. (C) Liver tissues from fasting Psmd10-transgenic mice or littermates were immunostained for PSMD10. Representative pictures are shown. Arrows indicate nuclear expression of PSMD10. (D) ChIP assay covering the region of the ATG7 promoter from −1809 to −1412 was performed to measure the binding activity of PSMD10 to the ATG7 promoter in HCC-LM3 cells after EBSS starvation for 12 or 24 h (upper panel). Quantification of the ratio of PSMD10 binding to IgG (lower panel). Data represent the mean ± SD of 3 independent experiments (**P < 0.01).
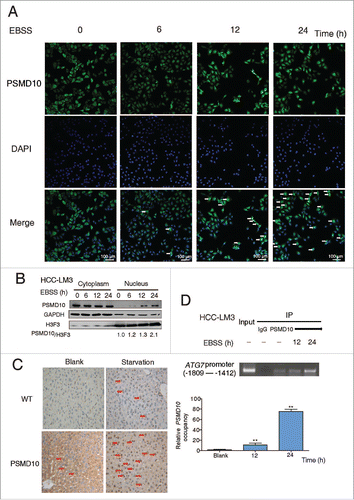
Figure 5. PSMD10 associates with HSF1 to cooperate in the expression of ATG7. (A) ChIP assay of the ATG7 promoter containing HSF1 binding sites from −1809 to −1412 was performed to measure the effect of PSMD10 on the binding activity of HSF1 under EBSS starvation for 24 h (left panel). Quantification of the ratio of HSF1 binding to IgG (lower panel). (B) The effect of PSMD10 on ATG7 reporter activity. SMMC-7721 cells were transiently cotransfected with pcDNA3.1/MYC-PSMD10 and ATG7 WT/mut1/mut2 luciferase plasmids. Luciferase activities were detected at 24 h after transfection and EBSS starvation. Data represent mean (±SD) from 3 independent experiments (*P < 0.05). (C) SMMC-7721 cells were transiently cotransfected with pcDNA3.1 or MYC-PSMD10 and a nonspecific RNAi control or siHSF1 RNA #3, and the indicated molecules were detected after EBSS starvation for 24 h. Data represent mean (±SD) from 3 independent experiments (*P < 0.05). (D) Cytoplasmic and nuclear fractionation from SMMC-7721-PSMD10 cells was performed for coimmunoprecipitation with MYC-tag antibody, followed by probing with anti-HSF1 or MYC-tag antibody. (E) HCC-LM3 cells treated with EBSS for 24 h were immunoprecipitated and immunoblotted for the indicated molecules. (F) HCC-LM3 cells were immunostained for endogenous PSMD10 and HSF1 to observe their colocalization in the presence or absence of EBSS for 24 h. DNA was stained by DAPI. Representative confocal microphotographs are shown together with profiles of colocalization within the area of interest (yellow signals in the nucleus).
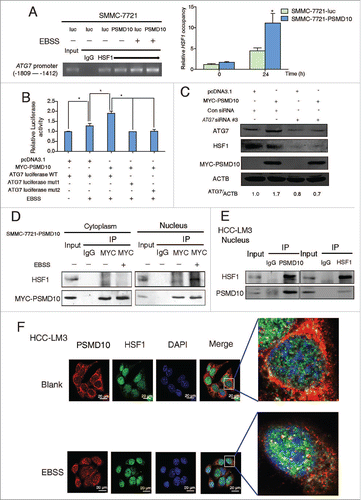
Figure 6. PSMD10 connects the proteasome and the autophagic pathway. (A) Proteasome activity and autophagy level were detected in SMMC-7721 with or without EBSS for the indicated time. (B) Proteasome activity was measured in the indicated cells treated with EBSS at the indicated times. (C) Expression of LC3B-II was detected after EBSS treatment combined with or without proteasome inhibitors (MG132 10 μM, ALLN 10 μM) for 6 or 12 h in SMMC-7721 and HCC-LM3 cells. (D) SMMC-7721-PSMD10 cells were treated as in (C), immunoprecipitated with MYC-tag antibodies, and then immunoblotted with anti-ATG7 or anti-MYC-tag antibodies. (E) Localization of PSMD10 in HCC-LM3 cells treated as in (C) was detected. (F) ChIP assay of the binding capability of PSMD10 to the ATG7 promoter in HCC-LM3 cells treated as in (C).
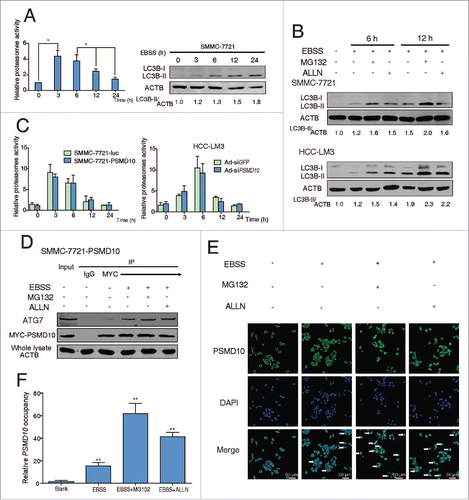
Figure 7. The relationship between PSMD10 and ATG7 in the clinical samples. (A, B) The liver tumor frozen sections from 2 patients were performed by immunofluorescence analysis to detect the localization of PSMD10 and ATG7 (A) or HSF1 (B). DNA was stained by DAPI. Representative confocal microphotographs are shown together with profiles of colocalization within the area of interest (yellow signals merge), and higher-magnification images of the outlined areas are also shown. (C) Proteins were extracted from 100 patients with HCC for immunoblot with anti-PSMD10 or anti-ATG7 antibody. The protein bands were analyzed by ImageJ2x software and the relative band intensity was normalized to ACTB band. Representative samples are shown (left panel). Correlation of expression level of PSMD10 and ATG7 is shown (right panel). (D) Representative PSMD10 and ATG7 expression in TMA sections of 117 patients with HCC (original magnification 200 ×; left panel). The median value for PSMD10 and ATG7 expression in HCCs was used to divide the patients into high (above median) and low (below median) PSMD10 and ATG7 groups. The disease-free and overall survival rates were compared between the low PSMD10 and ATG7 expression group (n = 28) and the high PSMD10 and ATG7 expression group (n = 27; right panel).
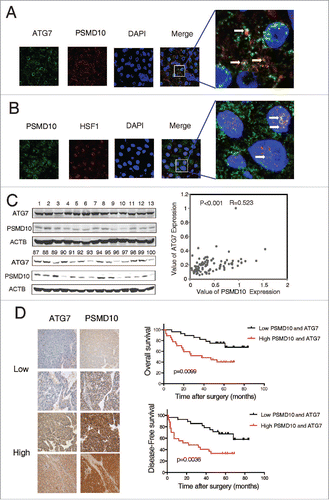
Figure 8. PSMD10 resisted apoptosis and enhanced autophagy under sorafenib treatment. (A) Representative PSMD10 expression in TMAs from 70 patients with HCC who underwent liver resection and sorafenib therapy (upper panel). Overall survival rates were compared between the low-PSMD10 (below the median value, n = 35) and high-PSMD10 groups (above the median value, n = 35) (lower panel). (B) SMMC-7721-luc or SMMC-7721-PSMD10 cells were treated with sorafenib (20 μM) for the indicated time or with the indicated dosage for 24 h. (C) LC3B-II was immunoblotted in SMMC-7721 or HCC-LM3 cells after sorafenib (20 μM) treatment for the indicated time. (D) The formation of LC3 puncta was detected in the indicated cells exposed to 20 μM sorafenib for 12 or 24 h. Quantification of the ratio of LC3 punctate cells to overall cells was shown in right panel. (E) Electron microscopy analysis showed autophagic vesicles or autophagosomes in cells from (D).
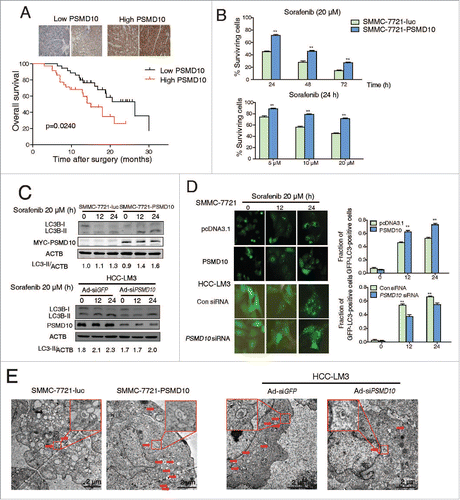
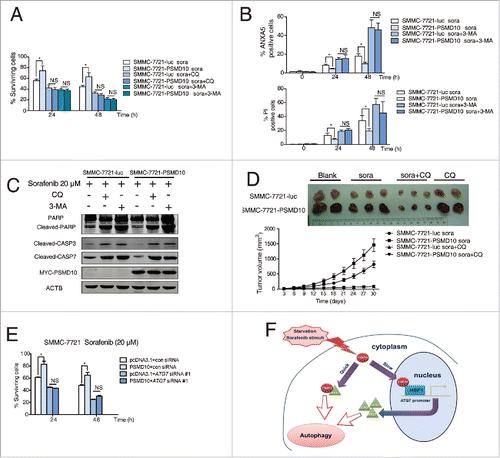
Figure 9. PSMD10-mediated autophagy caused drug resistance to sorafenib. (A) SMMC-7721-luc, SMMC-7721-PSMD10 cells were subjected to sorafenib (20 μM) for 24 or 48 h in the presence or absence of either CQ (10 μM) or 3-MA (10 mM) before performing CCK-8 assays. (B) Cells from (A) were analyzed for apoptosis. The percentage of cells was determined using the ANXA5 and propidium iodide (PtdIns) staining assay. (C) Immunoblot for apoptosis markers including PARP, CASP3, and CASP7 in cells from (B). (D) Effects of sorafenib and CQ on subcutaneous PSMD10-overexpressing tumors. SMMC-7721-luc or SMMC-7721-PSMD10 cells were injected into the right flanks of nude mice. Two weeks after injection, mice were randomly assigned into 4 groups, and set for sorafenib or CQ alone or both combination treatments (intraperitoneally, i.p.). Tumors from mice in each group, after tumor therapy, are presented, and average tumor volume for each group is shown. (E) SMMC-7721 cells were transiently cotransfected with pcDNA3.1 or MYC-PSMD10 and a nonspecific RNAi control or siATG7 RNA #1, then CCK-8 assays were performed. (F) A model for PSMD10, as a recyclable molecule to modulate autophagy in 2 different and complementary mechanisms. Upon the stressed condition, PSMD10 will first take part in an autophagic vacuole-forming process, through an interaction with cytosolic ATG7. Subsequently, PSMD10 will translocate into the nucleus and interact with HSF1 bound onto the ATG7 promoter to upregulate ATG7 expression, leading to further increased autophagy. Data represent mean (±SD) from 3 independent experiments (*P < 0.05).