Figures & data
Figure 1. Metabolic profile of male Atg7 cKO mice. (A) PCR using DNA from peritoneal Mφs and primers specific for Atg7. (B) Peritoneal Mφs were cultured in serum-free conditions for 6 h to induce autophagy, and cell extracts were subjected to western blot analysis. (C) Quantitative RT-PCR was done using mRNA from Mφs and specific primers. (D) Nonfasting blood glucose level was determined in male Atg7 cKO, Atg7 cKO-ob/ob and respective control mice (n = 10 to 15). (E) IPGTT was conducted in overnight-fasted 16-wk-old male mice as described in Materials and Methods, and AUC calculated (n = 5 to 8). (F) ITT was conducted in fasted 16-wk-old male mice as described in the Materials and Methods, and AUC calculated (n = 5 to 10). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
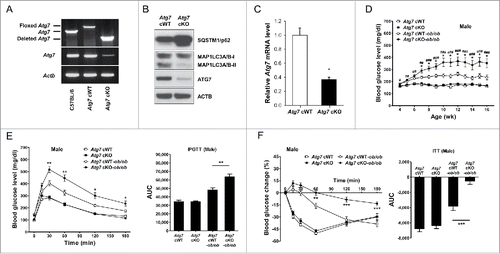
Figure 2. Inflammasome activation in Mφs from 12-wk-old mice by varying doses of PA in combination with 500 ng/ml LPS. (A) Primary peritoneal Mφs were incubated with PA of the indicated concentrations and LPS. After 24 h of culture, IL1B concentration in the supernatant was determined by ELISA. (B) After incubation of primary peritoneal Mφs with PA of the indicated doses and LPS for 24 h, cell extracts were subjected to western blot analysis using anti-IL1B and -CASP1 Abs. (C) After the same incubation of Mφs as in (B), NAD+:NADH ratio was determined using a commercial kit. (D) After the same incubation of Mφs as in (B), cells were incubated with MitoSOX Red for flow cytometry (dark green, unstained; green, Atg7 cWT Mφs treated with 400 μM PA; blue, Atg7 cWT Mφs treated with LPS; orange, Atg7 cWT Mφs treated with 400 μM PA + LPS; red, Atg7 cKO Mφs treated with 400 μM PA + LPS). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
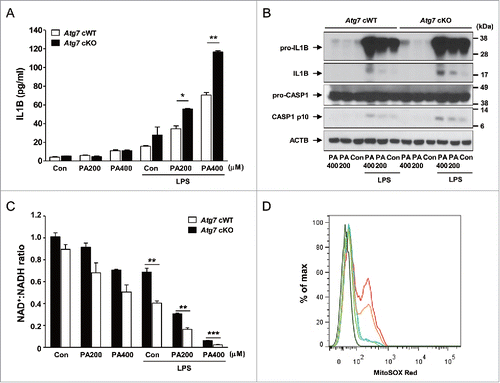
Figure 3. Inflammasome activation in vivo. (A) The number of CLS in white adipose tissue of 20-wk-old mice was counted as described in Materials and Methods (n = 4 each) (right). Representative H&E stained sections of white adipose tissue are shown (left, scale bar: 100 μm). (B) Total mRNA was extracted from the SVF of white adipose tissue from 20-wk-old mice, and quantitative RT-PCR was done using primers specific for Tnf, Il6, pro-Il1b and Adgre1 (n = 3 each). (C) Tissue lysate of the SVF was subjected to western blot analysis using anti-IL1B and -CASP1 Abs. (D) Serum IL1B level was determined using a commercial ELISA kit (n = 3 to 15). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
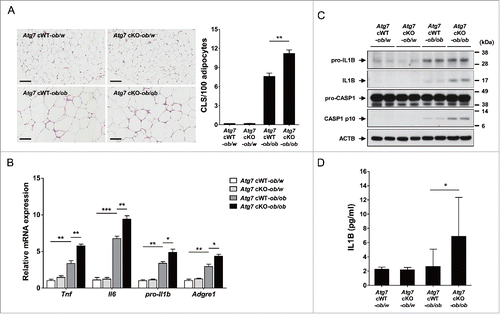
Figure 4. Treg cells in white adipose tissue. (A) Cells in the SVF of white adipose tissue from 20-wk-old mice were incubated with a mixture of anti-CD3E, -CD4, and -IL2RA Abs, and then with -FOXP3 Ab as described in Materials and Methods. FOXP3 and CD4 expression was examined by flow cytometry gated on CD3E+ CD4+ cells. The proportions of FOXP3+ cells among CD3E+ CD4+ cells (lower left) and the numbers of FOXP3+ CD3E+ CD4+ cells/gm adipose tissue (lower right) (n = 3 each). Representative FACS scattergrams are shown (upper). (B) Cells were incubated with Abs as in (A), and FOXP3 and IL2RA expression was examined by flow cytometry gated on CD3E+ CD4+ cells. The proportions of FOXP3+ IL2RA+ cells among CD3E+ CD4+ cells (lower left) and the numbers of FOXP3+ IL2RA+ CD3E+ CD4+ cells/gm adipose tissue (lower right) (n = 3 each). Representative FACS scattergrams are shown (upper). *, P < 0.05; **, P < 0.01.
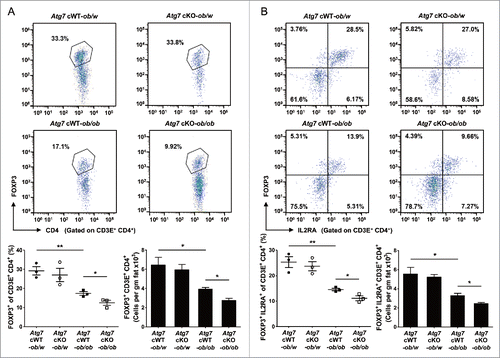
Figure 5. DSS-induced colitis in Atg7 cKO mice. (A) Body weight was monitored during and after oral administration of 2.5% DSS for 5 d (n = 7 each). The antibiotic mixture (Abx) was administered since 2 d before DSS as described in Materials and Methods, and body weight monitored (n = 7 each). (B) Gross morphology (upper) and length (lower) of the colons on day 7 of DSS administration (n = 5 to 6). (C) Survival after administration of 4% DSS was monitored as described in Materials and Methods (n = 10 each). *, P < 0.05; **, P < 0.01.
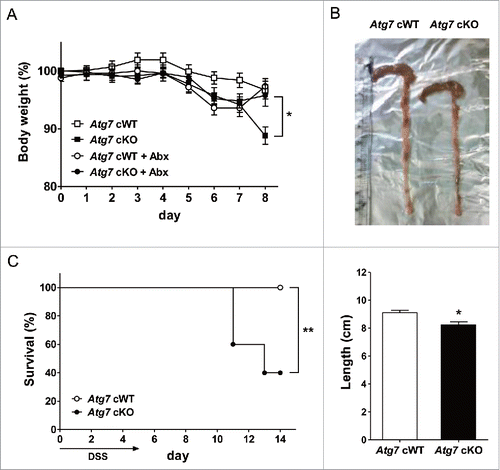
Figure 6. Inflammation and Th1 skewing in the colon of Atg7 cKO mice. (A) Colitis score was calculated on d 7 of DSS treatment as described in Materials and Methods (n = 5 each) (right). Representative H&E sections are shown (left, scale bar: 200 μm) (asterisk, inflammation; arrow, ulceration; arrow head, regeneration; white arrow, transmural lesion). (B) Relative expression of cytokines and chemokines in the colonic tissues on d 7 of DSS treatment assessed by quantitative RT-PCR analysis of mRNA prepared using a formalin-fixed paraffin-embedded extraction kit (n = 8 to 10). (C) Th skewing in the colonic lamina propria on day 7 of DSS treatment was evaluated by flow cytometry. The proportions of IFNG+ cells and IL17A+ cells among CD3E+ CD4+ T cells (lower) (n = 4 each). Representative scattergrams are shown (upper). *, P < 0.05.
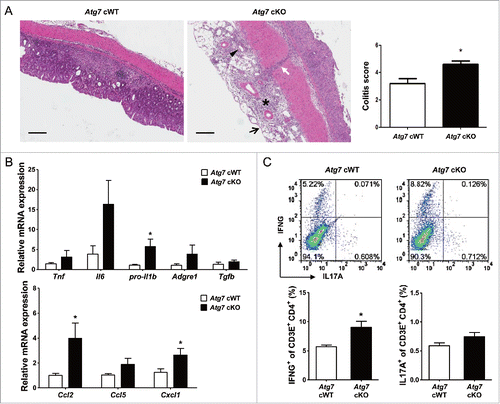
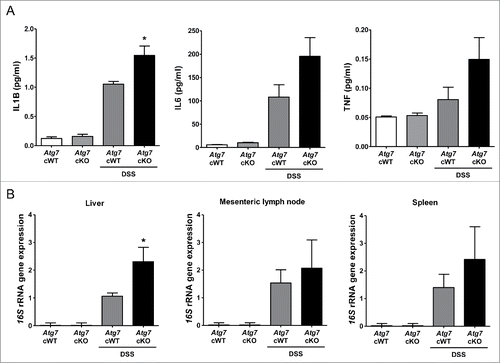
Figure 7. Systemic inflammation and bacterial invasion after DSS treatment. (A) Serum levels of cytokines after DSS treatment were determined by ELISA (n = 6 each). (B) Relative expression of bacterial 16S rRNA genes in the liver, mesenteric lymph node and spleen determined by quantitative PCR as described in Materials and Methods (n = 9 each). *, P < 0.05.