Figures & data
Figure 1. PLK1 binds and phosphorylates MTORC1, and PLK1 inhibition activates MTORC1 in interphase cells. (A) HeLa cells were cultured in full medium. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 4 independent experiments. (B) HeLa cells were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min and treated with the PLK1 inhibitor BI2536 for 30 min, as indicated. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (C) Quantification of data shown in (B). Ratio of RPS6KB (p70) phospho-(T389)/RPS6KB (p70) was calculated for n = 3 independent experiments. Data are normalized to 1 for the amino acid- and insulin-stimulated control condition, and represented as mean ± SEM. A one-way ANOVA followed by the Bonferroni multiple comparison test was applied; ns, nonsignificant; **, P ≤ 0.01. (D) PLK1 (shPLK1) or control (shControl) shRNA HeLa cells were starved for amino acids and growth factors for 1 h, and stimulated with amino acids and insulin for 30 min. Samples were analyzed by immunoblotting without removal of the mitotic cells. Data shown are representative of n = 3 independent experiments. (E) Quantification of data shown in (D). Ratio of RPS6KB (p70) phospho-(T389):RPS6KB (p70) was calculated for n = 3 independent experiments. Data are normalized to 1 for the amino acid- and insulin-stimulated shControl condition, and represented as mean ± SEM. A one-way ANOVA followed by the Bonferroni multiple comparison test was applied; ns, nonsignificant. (F) RPTOR shRNA (shRPTOR) or shControl HeLa cells were arrested in mitosis by nocodazole treatment. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (G) PLK1 (shPLK1) or control (shControl) shRNA HeLa cells were starved for amino acids and growth factors for 16 h and stimulated with amino acids and insulin for 35 min. Mitotic cells were removed by shake-off. Samples were analyzed by immunoblotting. Data are representative of n = 4 independent experiments. (H) Quantification of data shown in (G). Ratio of RPS6KB (p70) phospho-(T389):RPS6KB (p70) was calculated for n = 4 independent experiments. Data are normalized to 1 for the amino acid- and insulin-stimulated shControl condition and represented as mean ± SEM. A one-way ANOVA followed by the Bonferroni multiple comparison test was applied; ns, nonsignificant; **, P ≤ 0.01. (I) HeLa cells were treated with BI2536 and/or Torin1 as indicated, and stimulated as described in (B). Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (J) Quantification of data shown in (I). Ratio of RPS6KB (p70) phospho-(T389):RPS6KB (p70) was calculated for n = 3 independent experiments. Data are normalized to 1 for control condition (no Torin1, no BI2536), and represented as mean ± SEM. A one-way ANOVA followed by the Bonferroni multiple comparison test was applied; ns, nonsignificant; **, P ≤ 0.01. (K) PLK1 kinase assay. HA-RPTOR was immunopurified from HeLa cells. An unspecific IgG antibody was used as negative control. All samples were dephosphorylated before adding them to the kinase reaction with recombinant PLK1. Data shown are representative of n = 3 independent experiments. IP, immunoprecipitation; IB, immunoblot; KA, kinase assay. (L) Quantification of data shown in (K) for n = 3 independent experiments. Data are normalized to 1 for HA-RPTOR phosphorylation by PLK1. Data are represented as mean ± SEM. A one-way ANOVA followed by the Bonferroni multiple comparison test was applied; ns, nonsignificant; **, P ≤ 0.01. (B, C, D, E, G, H, I) aa, amino acids; ins, insulin.
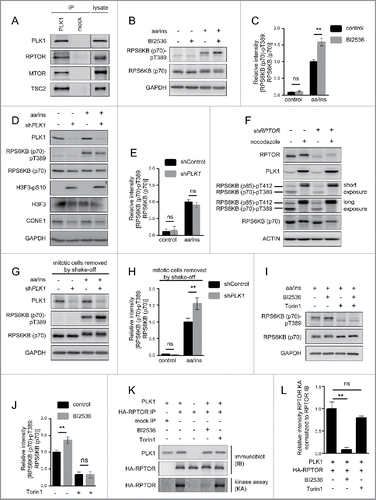
Figure 2. PLK1 resides with MTORC1 at lysosomes, and overexpression of active PLK1 decreases lysosomal association of the PLK1-MTORC1 complex. (A) Immunofluorescence analysis of HeLa cells that were cultured in full medium and stained with LAMP2 and MTOR antibodies. White regions in merged image (right) of LAMP2 (green) and MTOR (magenta) indicate colocalization. Nuclei were stained with Hoechst 33342. Scale bar 20 µm. Representative images are shown for n = 3 independent experiments. (B) Immunofluorescence analysis of HeLa cells that were cultured in full medium and stained with PLK1 and MTOR antibodies. White regions in merged image (right) of PLK1 (green) and MTOR (magenta) indicate colocalization. Nuclei were stained with Hoechst 33342. Scale bar 20 µm. Representative images are shown for n = 3 independent experiments. (C) Immunofluorescence analysis of HeLa cells that were synchronized in prometaphase with nocodazole for 16 h and released for 30 min in full medium. Cells were stained with PLK1 antibody. Nuclei were stained with Hoechst 33342. Scale bar: 10 µm. Representative images of cells in metaphase (left) and anaphase (right) are shown for n = 3 independent experiments. (D) Analysis of input sample taken before fractionation in sucrose gradient (E). The mitotic cell lysate was collected from HeLa shPLK1 knockdown cultures without mitotic shake-off. Samples were analyzed by immunoblotting. Data shown are representative of n = 2 independent experiments. (E) HeLa cells were starved for 1 h for amino acids and growth factors and stimulated with amino acids and insulin for 35 min. Samples were separated in a 10 to 40% sucrose gradient and analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (F) Quantification of data shown in (E) for n = 3 independent experiments. The percentage of PLK1 in either the lysosomal or the nuclear fraction is displayed. Data are represented as mean ± SEM. (G) HeLa cells were cultured in full medium. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (H) HeLa cells overexpressing wild type MYC-PLK1 (WT) or empty vector were cultured in full medium. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (I) HeLa cells overexpressing MYC-PLK1 (WT) or kinase-defective, dominant negative MYC-PLK1K82R were cultured in full medium. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (J) HeLa cells overexpressing MYC-PLK1 (WT) or kinase-defective, dominant negative MYC-PLK1K82R were starved for 1 h for amino acids and growth factors, and stimulated with amino acids and insulin for 35 min. Cells were then starved for amino acids for 10 min as indicated, and samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments.
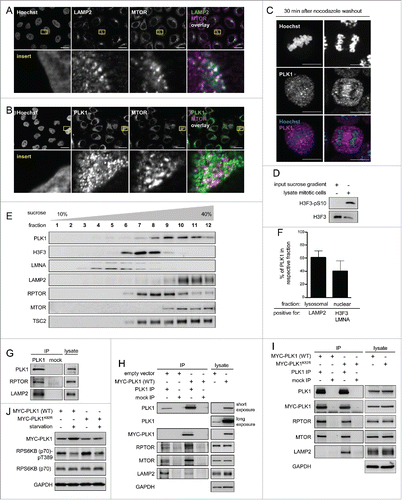
Figure 3. PLK1 inhibition hyperactivates MTORC1 and increases lysosomal MTORC1 localization in amino acid-starved interphase cells. (A) HeLa cells were starved for 1 h for amino acids and growth factors, and stimulated with amino acids and insulin for 35 min. Cells were then starved for amino acids for 5, 10, 15 and 30 min and treated with BI2536 or carrier, as indicated. Samples were analyzed by immunoblotting. Data shown are representative of n = 4 independent experiments. (B) Quantification of data shown in (A). Ratio of RPS6KB (p70) phospho-(T389):RPS6KB (p70) was calculated for n = 4 (5 min starvation and 15 min starvation); n = 3 (10 min starvation) independent experiments. Data are normalized to 1 for starvation control condition and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (C) Immunofluorescence analysis of HeLa cells that were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min, followed by 30 min amino acid starvation, without or with the PLK1 inhibitor BI2536. Staining was performed with MTOR and LAMP2 antibodies. White regions in merged image (right) of MTOR (green) and LAMP2 (magenta) indicate colocalization. Nuclei were stained with Hoechst 33342. Scale bar 20 µm. Representative images are shown for n = 3 independent experiments. (D) Analysis of MTOR-LAMP2 colocalization by the Pearson correlation coefficient for experiment shown in (C). Data are represented as mean ± SEM, and are representative of n = 3 independent experiments. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (E) Immunofluorescence analysis of HeLa cells that were treated as described in (C). Staining was performed with RRAGC and LAMP2 antibodies. White regions in merged image (right) of RRAGC (green) and LAMP2 (magenta) indicate colocalization. Nuclei were stained with Hoechst 33342. Scale bar 20 µm. Representative images are shown for n = 3 independent experiments. (F) Analysis of RRAGC-LAMP2 colocalization by the Pearson correlation coefficient for experiment shown in (E). Data are represented as mean ± SEM, and are representative of n = 3 independent experiments. A nonparametric 2-tailed Student t test was applied; ns, nonsignificant. (G) HeLa cells were either cultured in full medium or starved for amino acids and growth factors for 16 h. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (H) Quantification of IP samples shown in (G). Ratio of MTOR:PLK1 was calculated for n = 3 independent experiments. Data are normalized to 1 for control condition and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (I) HeLa cells were treated as described in (G). IP was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 4 independent experiments. (J) Quantification of IP samples shown in (I). Ratio of RPTOR:PLK1 was calculated for n = 4 independent experiments. Data are normalized to 1 for control condition and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05.
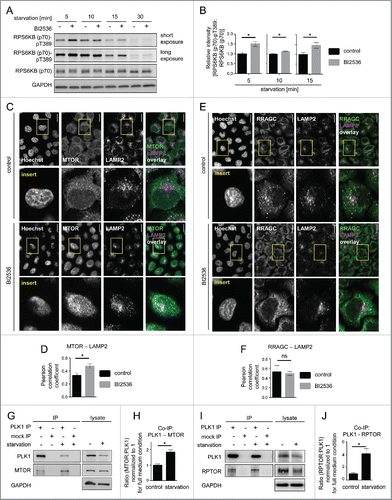
Figure 4. Starvation enhances PLK1-MTOR binding and cytoplasmic PLK1-MTOR colocalization. (A) HeLa cells were cultured in full medium and treated for 30 min with Torin1 or carrier, respectively. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (B) Quantification of IP samples shown in (A). Ratio of MTOR:PLK1 was calculated for n = 3 independent experiments. Data are normalized to 1 for full medium condition and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; ns, nonsignificant. (C) HeLa cells were either cultured in full medium or starved for amino acids and growth factors for 16 h. Cells were then treated with BI2536 or carrier, as indicated. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (D) Quantification of data shown in (C). Fold change of MTOR:PLK1 ratio in starved versus control cells was calculated across n = 3 independent experiments, for carrier or BI2536 treated cells, as indicated. Data are normalized to lane 1 (C), and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; ns, nonsignificant. (E) HeLa cells were starved for 1 h for amino acids and growth factors, and stimulated with amino acids and insulin for 35 min. Afterwards, for starvation, amino acids were withdrawn for 30 min. Immunoprecipitation (IP) was performed with PLK1 and control (mock) antibodies. Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (F) Quantification of data shown in (E). Ratio of MTOR:PLK1 was calculated for n = 3 independent experiments. Data are normalized to 1 for amino acids and insulin condition, and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; ***, P ≤ 0.001. (G) Immunofluorescence analysis of HeLa cells that were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min, followed by 30 min of amino acid starvation, as indicated. Staining was performed with PLK1 and MTOR antibodies. White regions in merged image (right) of PLK1 (green) and MTOR (magenta) staining indicate colocalization; insert 1 shows a region with lysosomal MTOR pattern; insert 2 shows a cytoplasmic region without lysosomal MTOR pattern. Nuclei were stained with Hoechst 33342. Scale bar: 20 µm. Representative images are shown for n = 3 independent experiments. (H) Analysis of PLK1-MTOR colocalization by the Pearson correlation coefficient for experiment shown in (G). Ten representative cells were quantified for each condition. Data are represented as mean ± SEM and are representative of n = 3 independent experiments. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (E, F, G, H) aa, amino acids; ins, insulin.
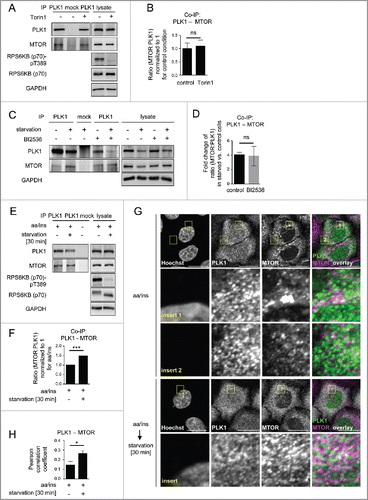
Figure 5. PLK1 inhibition reduces the autophagy marker LC3-II in interphase cells. (A) HeLa cells were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min, followed by 30 min amino acid starvation. All media were supplemented with bafilomycin A1. BI2536 was added as indicated for 30 min. Data shown are representative of n = 3 independent experiments. (B) Quantification of data shown in (A). Ratio of LC3-II:GAPDH was calculated for n = 3 independent experiments. Data are normalized to 1 for the control condition and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (C) HeLa cells were treated with BI2536 and/or Torin1 as indicated, and stimulated as described in (A). Samples were analyzed by immunoblotting. Data shown are representative of n = 3 independent experiments. (D) Quantification of data shown in (C). Ratio of LC3-II:GAPDH was calculated for n = 3 independent experiments. Data are normalized to 1 for the control condition (no Torin1, no BI2536) and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; ns, nonsignificant; *, P ≤ 0.05. (E) PLK1 (shPLK1) or control (shControl) shRNA HeLa cells were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min, followed by 20 min amino acid starvation. All media were supplemented with bafilomycin A1. Cells were treated with Torin1 as indicated. Mitotic cells were removed by shake-off. Hence, only interphase cells were analyzed. Data shown are representative of n = 4 independent experiments. (F) Quantification of data shown in (E). Ratio of LC3-II:GAPDH was calculated for n = 4 independent experiments. Data are normalized to 1 for the shControl condition (no Torin1) and represented as mean ± SEM. A nonparametric 2-tailed Student t test was applied; ns, nonsignificant; *, P ≤ 0.05.
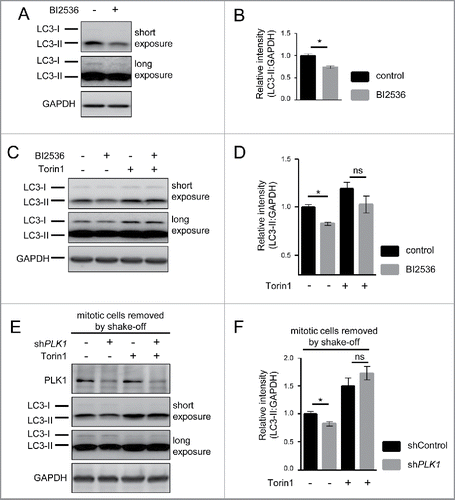
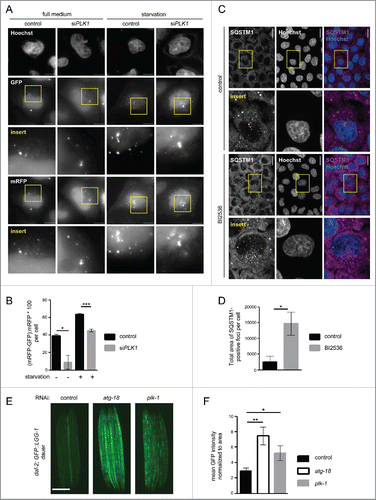
Figure 6. PLK1 inhibition impairs autophagy in nonmitotic cells and in C. elegans dauers. (A) HeLa cells were transfected with mRFP-GFP-LC3 tandem reporter, followed by PLK1 siRNA transfection on the next day. Cells were either kept in full medium, or starved for serum and amino acids for 16 h. Mitotic cells were removed by shake-off before fixation of cells 24 h after siRNA transfection. Representative images are shown for each condition. Scale bar 10 µm. Data shown are representative of n = 2 independent experiments. (B) Quantification of experiment shown in (A). The numbers of green puncta (autophagosomes) and red puncta (autolysosomes plus autophagosomes) were counted for nonmitotic cells. Data shown are represented as mean ± SEM for n = 30 cells for control knockdown, full medium, n = 43 cells for siPLK1, full medium, n = 35 cells for control knockdown, starvation condition, and n = 26 for siPLK1 starvation condition. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05; ***, P ≤ 0.001. (C) Immunofluorescence analysis of HeLa cells that were starved for 1 h for amino acids and growth factors, stimulated with amino acids and insulin for 35 min, followed by 30 min amino acid starvation. All media were supplemented with bafilomycin A1. Staining was performed with SQSTM1 antibody and Hoechst 33342. Shown are maximum intensity projections. Scale bar 20 µm. Representative images are shown for n = 3 independent experiments. (D) Quantification of experiment shown in (C). The total area of SQSTM1-positive foci was calculated and normalized to the number of nuclei. n = 126 cells for control condition and n = 105 cells for BI2536 treated condition. Data are represented as mean ± SEM, and are representative of n = 3 independent experiments. A nonparametric 2-tailed Student t test was applied; *, P ≤ 0.05. (E& F) daf-2(e1370) animals expressing GFP::LGG-1 were fed bacteria expressing control, atg-18 or plk-1 dsRNA from hatching, and raised at the nonpermissible temperature (25°C) to induce dauers. (E) Micrographs of ∼8 to 10 dauer animals lined up next to each other were taken 6 d after the temperature shift. Scale bar 200 µm. (F) GFP::LGG-1 fluorescence (mean ± s.d. of ∼8–10 animals, **P<0.0001, one-way ANOVA) was quantified. Data shown are representative of 3 independent experiments.