Figures & data
Figure 1. Starvation promotes LDs formation in the basolateral side of kidney proximal tubules. LDs formation and its localization in the kidney were investigated in wild-type mice. (A and B) Images of Oil Red O- (A) and toluidine blue (B) -stained kidney sections of fed and 12- or 24-h starved mice (n = 6 to 9 in each group). Counterstaining was performed with hematoxylin (A). (C) Images of staining for COL4A4 (red) and BODIPY 493/503 (green) in 24-h starved mice. (D to K) Electron microscopy images of fed (D to G) and 24-h starved mice (H to K) (n = 3 in each group). BM, basement membrane; TL, tubular lumen; Mt, mitochondria; LD, lipid droplet; *, nucleus. (L) Three-dimensional reconstruction images of immunofluorescence staining for COX4I1 (red) and BODIPY 493/503 (green) in 24-h starved mice. Bars: 50 μm (A and C), 20 μm (B and L), 10 μm (H), 5 μm (I), and 2 μm (J and K). All images are representative of multiple experiments.
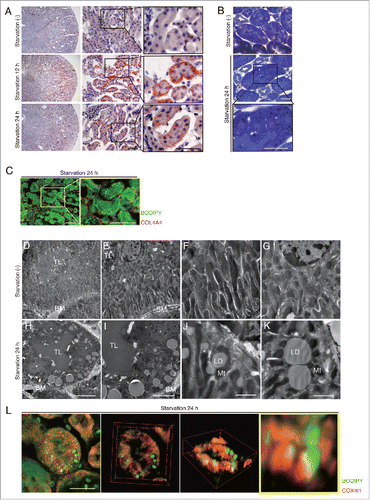
Figure 2. Starvation-induced autophagic flux peaks at 24 h in PTCs. (A and B) Autophagic flux was assessed in the proximal tubules of GFP-MAP1LC3B transgenic that were either fed or subjected to 24 or 48 h of starvation, with or without chloroquine administration 6 h before euthanasia (n = 6 to 9 in each group). Kidney sections were immunostained for LRP2/MEGALIN, a marker of proximal tubules (red), and counterstained with DAPI (blue). Arrows indicate GFP- MAP1LC3B-positive puncta. (B) The number of GFP-positive puncta per proximal tubule under each condition was counted in at least 10 high-power fields (× 600). (C) Western blot analysis using whole kidney lysates of wild-type mice that were fed or subjected to 24 or 48 h of starvation with or without chloroquine administration (n = 5 to 6 in each group). Quantification by densitometry in protein levels of MAP1LC3-II is shown. Data are expressed as the fold change relative to the mean value of fed control mice without chloroquine administration. (D) Images of staining for protein aggregates detected by ProteoStat dye in the kidney of fed and 24- or 48-h starved mice with or without chloroquine administration 6 h before euthanasia (n = 3 to 4 in each group). Counterstaining was performed with Hoechst 33342 (blue). Arrows indicate protein aggregates. (E) The number of protein aggregates per proximal tubule under each condition was counted in at least 10 high-power fields (× 600). Bars: 20 μm. Images are representative of multiple experiments. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05) are indicated. n.s., not significant.
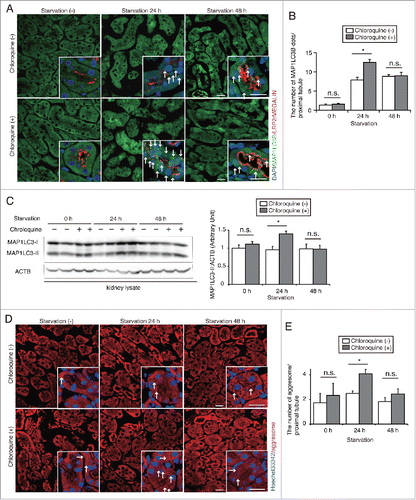
Figure 3. Prolonged starvation induces lipophagy in the kidney proximal tubules. Lipophagy in the PTCs was investigated immunohistochemically, morphometrically and biochemically. (A and B) Colocalization of LDs and autophagosomes was assessed in the proximal tubules of fed and 24- or 48-h starved mice, with or without chloroquine administration 6 h before euthanasia (n = 5 in each group). (A) Kidney sections were stained for MAP1LC3B (red) and BODIPY 493/503 (green), and counterstained with DAPI (blue). Arrows indicate MAP1LC3B dots colocalized with BODIPY 493/503. (B) The number of BODIPY and MAP1LC3B-merged dots per proximal tubule under each condition was counted in at least 10 high-power fields (× 600). Data are provided as mean ± SE. Statistically significant differences (*P < 0.05) are indicated. n.s., not significant. (C to F) Electron microscopy analysis of kidney sections in 48-h starved mice. Arrows indicate double-membrane autophagic vacuoles containing only lipids (C to E) and elongating phagophores along the surface of cytosolic LDs (F) (n = 3 in each group). (E and F) Magnified images are presented in the insets. BM, basement membrane; TL, tubular lumen; Mt, mitochondria; LD, lipid droplet. Bars: 10 μm (A), 5 μm (C and D) and 500 nm (E and F). (G) Immunoblots images using homogenates (HOM) and isolated LDs from the kidneys of fed and 24- or 48-h starved mice (n = 5 to 6 in each group). All images are representative of multiple experiments.
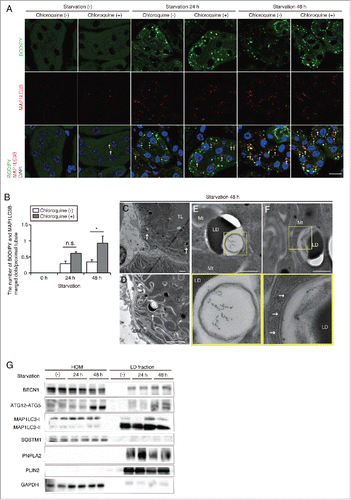
Figure 4. Lipophagy mobilizes FAs from LDs to mitochondria during starvation in vitro. FA mobilization following lipophagy in starved cultured PTCs was investigated by a pulse-chase assay using Red C12, fluorescent FA analog. (A) Schematic representation of the pulse-chase assay. PTCs stably expressing GFP-MAP1LC3B were supplied with 250 μM OA and 1 μM of Red C12 for 12 h, and chased with serum-limited medium (0.1% FBS) for the indicated periods of time and the subcellular localization of Red C12 was determined. Lysosomes were stained with LAMP1 (white) and counterstained with DAPI (blue) (n = 5 in each group). (B) A bar graph of Red C12 localization is shown. The following groups of Red C12 are expressed as percentage of total Red C12 area for each condition: Red C12 negative for GFP-MAP1LC3B and LAMP1, Red C12 positive for GFP-MAP1LC3B or LAMP1, and Red C12 positive for both GFP-MAP1LC3B and LAMP1. (C) OA- and Red C12-treated autophagy-competent or -deficient PTCs with or without 24 h of starvation with serum-limited medium (0.1% FBS) were stained with MitoTracker Green FM (n = 5 in each group). Colocalization of Red C12 and mitochondria was assessed by the Pearson correlation. Bars: 10 μm. All images are representative of multiple experiments. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05) are indicated. n.s., not significant. Atg5+, autophagy-competent PTC; atg5−/−, autophagy-deficient PTC.
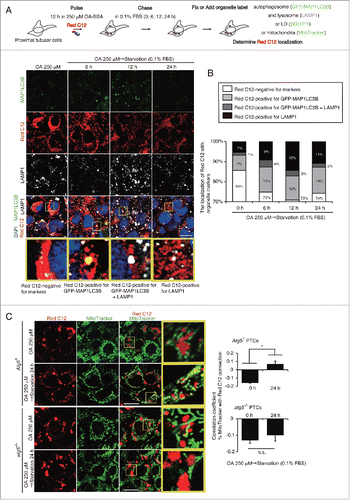
Figure 5. Lipophagy produces ATP for cell survival in the proximal tubules during starvation in vitro. Effects of starvation-induced lipophagy on cellular energy homeostasis and cell survival were studied. (A and B) Autophagy-competent and -deficient PTCs were supplied with 250 μM OA containing Red C12 for 12 h and then incubated with serum-limited medium (0.1% FBS) for the indicated periods of time. LDs were labeled with BODIPY 493/503. BODIPY 493/503-positive LD content per cell was quantified in the indicated conditions. Counterstaining was performed with DAPI (blue) (n = 5 in each group). (C to E) Autophagy-competent and -deficient PTCs were supplied with 250 μM OA for 12 h and then incubated with HBSS with or without 40 μM etomoxir for the indicated periods of time (n = 3 to 5 in each group). (C) Images of Oil Red O staining. (D and E) Cellular ATP level (D) and cell survival rate (MTS assay) (E) were measured. (B and E) Data are expressed as the fold change relative to the mean value of control PTCs. Bars: 10 μm (A) and 50 μm (C). All images are representative of multiple experiments. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05 vs. autophagy-competent cells of the corresponding condition; #P < 0.05 vs. control PTCs) are indicated. Atg5+, autophagy-competent PTC; atg5−/−, autophagy-deficient PTC.
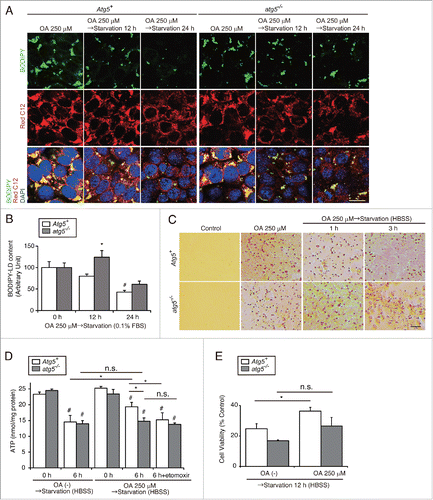
Figure 6. Autophagy-deficiency impairs LD degradation under prolonged starvation in the proximal tubules in vivo. Kidneys of atg5-TSKO and atg5F/F-CTRL mice, that were fed or starved for up to 48 h, were analyzed for the LD degradation (A to C, n = 10 to 14; D, n = 7 to 9; E, n = 5 to 6; F to I, n = 3 in each group). (A) Images of Oil Red O-stained kidney sections. Counterstaining was performed with hematoxylin. (B) Quantification of the Oil Red O staining. (C to E) TGs level per kidney weight (gKW) (C) and mRNA expression level of Plin2 in the kidney (D), immunoblots of the indicated molecules using homogenates (HOM) and isolated LD fraction from the whole kidney (E), and images of toluidine blue-stained kidney sections (F) are shown. (G to I) Transmission electron microscopy images of 48-h starved atg5-TSKO mice. BM, basement membrane. (D) Data are expressed as the fold change relative to the mean value of fed atg5F/F-CTRL mice. Bars: 50 μm (A), 10 μm (F and G), 5 μm (H), and 2 μm (I). All images are representative of multiple experiments. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05 vs. 48-h starved atg5F/F-CTRL mice; #P < 0.05 vs. fed mice) are indicated. CTRL, atg5F/F-CTRL mice; TSKO, atg5-TSKO mice.
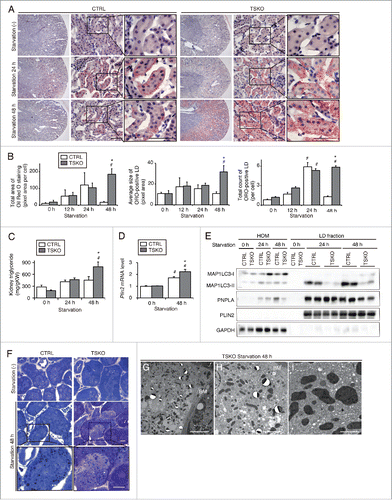
Figure 7. Impairment of lipid utilization induced by autophagy-deficiency under prolonged starvation is compensated by ketone bodies. Metabolic and energetic changes of atg5F/F-CTRL and atg5-TSKO mice, that were fed and starved for up to 48 h, were investigated using plasma and organs including kidney and liver (n = 7 to 9 in each group). (A) Representative images of immunostaining of PPARA in the kidney cortical regions. Kidney sections were immunostained for the proximal tubule marker LRP2/MEGALIN in blue. (B to E) mRNA expression levels of genes related to lipid β-oxidation (B) and to mitochondrial oxidative phosphorylation (C), ATP content per protein in the kidney (D), and plasma ketone body concentration (E) are shown. (F) mRNA expression level of Hmgcs2 in the liver. (G) Representative images of immunostaining for PPARΑ in the liver. Tissue sections were counterstained with hematoxylin. (H) mRNA expression levels of genes related to lipid β-oxidation in the liver. (B, C, F and H) Data are expressed as the fold change relative to the mean value of fed atg5F/F-CTRL mice. Cpt1a, carnitine palmitoyltransferase 1a, liver; Acads, acyl-Coenzyme A dehydrogenase, short-chain; Acadl, acyl-Coenzyme A dehydrogenase, long-chain; Acadvl, acyl-Coenzyme A dehydrogenase, very long chain. Bars: 50 μm (A) and 100 μm (G). All images are representative of multiple experiments. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05 vs. 48-h starved atg5F/F-CTRL mice; #P < 0.05 vs. fed mice) are indicated. n.s., not significant. CTRL, atg5F/F-CTRL mice; TSKO, atg5-TSKO mice.
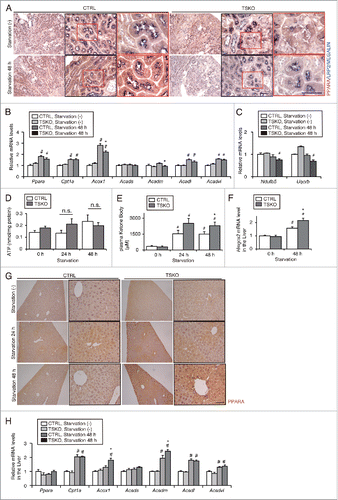
Table 1. Metabolic profiles in fed or starved atg5-TSKO and atg5F/F-CTRL mice.
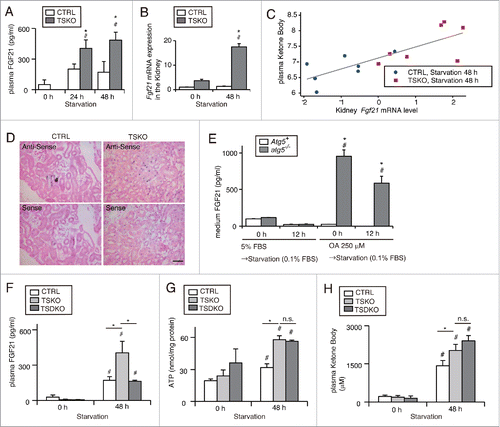
Figure 8. Increased secretion of FGF21 from autophagy-deficient kidney is associated with increased ketone bodies under prolonged starvation. FGF21 expression in the PTCs in response to autophagy-deficiency was investigated in vivo and in vitro. (A to C) Plasma concentration (A) and mRNA expression level in the kidney (B) of FGF21 in fed and 24- or 48-h starved atg5-TSKO and atg5F/F-CTRL mice (n = 10 to 14 (A), 7 to 9 (B) in each group). (B) Data are expressed as the fold change relative to the mean value of fed atg5F/F-CTRL mice. (C) Correlation between plasma ketone body concentration and Fgf21 mRNA expression level in the kidney on log-scale (n = 14; R2 = 0.622; P < 0.001, as determined by linear regression analysis). (D) Representative images of in situ hybridization of Fgf21 in the kidney of 48 h starved atg5F/F-CTRL and atg5-TSKO mice. Bars: 50 μm. CTRL, atg5F/F-CTRL mice; TSKO, atg5-TSKO mice. (E) Medium FGF21 concentration in autophagy-competent and -deficient PTCs that were incubated with 5% FBS or 250 μM OA for 12 h followed by starvation with 0.1% FBS for 12 h (n = 5 in each group). Atg5+, autophagy-competent PTC; atg5−/−, autophagy-deficient PTC. (F to H) Plasma FGF21 concentration (F), ATP per protein amount in the kidney (G) and plasma ketone body concentration (H) in atg5F/F or fgf21F/F-CTRL, Fgf21+/+atg5-TSKO and fgf21, atg5-TSKO mice under fed or 48-h starved condition (n = 5 in each group). CTRL, atg5F/F or fgf21F/F-CTRL mice; TSKO, Fgf21+/+atg5-TSKO mice; TSDKO, fgf21, atg5-TSKO mice. Data are provided as mean ± SE. Statistically significant differences (*P < 0.05 vs. atg5F/F-CTRL mice under the corresponding condition (A and B); *P < 0.05 vs. autophagy-competent cells of the corresponding condition (E); #P < 0.05 vs. fed mice (A and B, F to H); #P < 0.05 vs. control PTCs (E)) are indicated. n.s., not significant.