Figures & data
Figure 1. The n-3 PUFA DHA induces SQSTM1/p62-bodies in macrophages. (A) Confocal analysis of RAW264.7 cells treated with DHA, OA or AA (70 µM) for 8 h (scale bars: 10 µm) and (B) automated quantification of SQSTM1- and Ub-positive speckles (< 7000 cells/condition). Data are representative of 3 independent experiments of which 2 were manually counted. (C) Immunoblot (IB) analysis and quantification (below) of RAW264.7 cells treated with DHA, OA and AA (70 µM) for 16 h, n = 4, (repeated measures ANOVA with Dunnett's). (D) Confocal analysis of MDMs treated with DHA, OA or AA (70 µM) for 24 h (scale bars: 10 µm) and (E) automated quantification of SQSTM1/p62-bodies (< 4000 cells/condition), n = 3 except for 48 h where n = 1. (F) IB and quantification (below) of MDMs treated with DHA, OA and AA (70 µM) for 16 h, n = 5, (Friedman test with Dunn's).
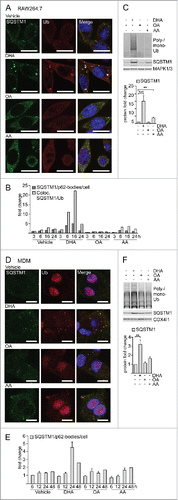
Figure 2. DHA induces formation of detergent-resistant SQSTM1/p62-bodies degraded by autophagy and activates NFE2L2. IB of soluble and insoluble fractions of RAW264.7 cells treated with DHA (70 µM) with or without BafA1 (100 nM) for (A) 16 h or (B) 8 h followed by 12 h incubation in fresh medium (Med) with or without BafA1. Hmw SQSTM1 was visualized by long exposure (exp). (C) IB of RAW264.7 cells treated with DHA, OA or AA (70 µM) with or without BafA1 (100 nM) for 8 h. Data are representative of 3 independent experiments. (D) FACS analysis of ROS levels in RAW264.7 cells at different time points after DHA (70 µM) using a fluorescent CM-H2DCFDA probe, n = 3, (paired t-test). (E) Confocal analysis of RAW264.7 cells treated with DHA, OA or AA (70 µM) for 3 h (scale bars: 10 µm). Representative images are shown, n = 3. (F) IB analysis of cytosolic (Cyt) or nuclear (Nuc) fractions after 3 h DHA with or without addition of 5 mM NAC or 6 h DHA and quantification of cytosolic SQSTM1 levels and nuclear NFE2L2 levels (below), n = 3, (repeated-measures ANOVA with Dunnett's). (G) IB analysis and quantification (right) of RAW264.7 cells after 3 h DHA with or without addition of 5 mM NAC, n = 3, (repeated-measures ANOVA with Dunnett's). (H) IB analysis and quantification (right) of MDM after 3 h DHA with or without addition of 5 mM NAC, n = 3. (I) QRT-PCR analysis of NFE2L2 target genes SQSTM1, HMOX1, NQO1 and SLC7A11 in vehicle- and DHA-treated MDMs, n = 5 (Friedman test with Dunn's).
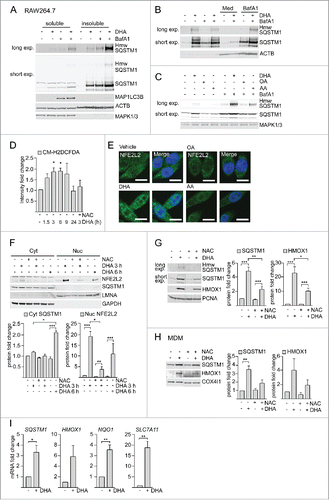
Figure 3. The n-3 PUFA DHA strongly reduced expression of CXCL10 by affecting several signaling pathways. (A) MDMs were treated with DHA (70 µM) for 16 h before stimulation with LPS (100 ng/ml) for 3 h. Transcript levels of 579 genes (nCounter GX Human Immunology kit assay) were determined. Transcripts that were more than 1.5-times increased by LPS in both donors were selected and the fold change between vehicle- and DHA-treated cells calculated, mean ± SD, n = 2. (B) QRT-PCR analysis of CXCL10 and (C) TNF in MDM treated with DHA, OA or AA for 16 h and LPS for 3 h, n = 5 (Friedman test with Dunn's). (D) IB analysis and quantification of IRF3 and RELA phosphorylation in MDMs treated with DHA, OA or AA for 16 h and LPS for 1 h, n = 5. (Friedman test with Dunn's), of (E) TBK1, IKBKB, STAT1 and STAT3 phosphorylation in MDMs treated with DHA, OA or AA for 16 h and LPS for 1 or 4 h, n = 5 (Wilcoxon matched-pairs signed rank test, one-tailed) and of (F) IRF1 and RELA in cytosolic (Cyt) and nuclear (Nuc) fractions in MDMs treated with DHA, OA or AA for 16 h and LPS for 2 or 4 h, n = 5 (Wilcoxon matched-pairs signed rank test). (G) IFNB1 mRNA levels after incubation with DHA for 16 h and LPS for 2 h, n = 6 (Wilcoxon matched-pairs signed rank test).
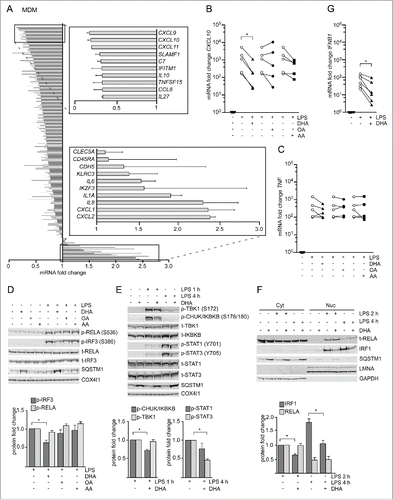
Figure 4. The DHA-mediated inflammatory tolerance is related to the formation of SQSTM1/p62-bodies. (A) IB analysis of MDMs treated with DHA (70 μM) for the indicated times followed by LPS for 1 h. The quantification (below) represents mean fold changes ± SEM of DHA + LPS compared to vehicle + LPS on a log scale, n = 6, (Friedman test with Dunn's). (B) QRT-PCR analysis of CXCL10 levels in MDM after 16 h DHA with and without addition of 5 mM NAC followed by 3 h LPS, n = 3, (repeated-measures ANOVA with Dunnett's). (C) QRT-PCR analysis of CXCL10 in SQSTM1 siRNA-transfected MDMs treated with DHA for 16 h and LPS for 3 h, n = 5, (Friedman test with Dunn's). (D) IB analysis of IRF3, RELA and IKBKB phosphorylation in control and SQSTM1 knockdown MDMs, n = 6 (Friedman test with Dunn's). (E) CXCL10 protein levels in Sqstm1+/+ (n = 8) and sqstm1−/− (n = 10) male mice were measured by ELISA. Serum was analyzed at baseline and at 3 and 6 h after challenge with 3.5 µg/g LPS. ND, not detected.
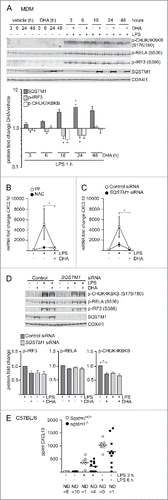
Figure 5. KEAP1 is sequestered in DHA-induced SQSTM1/p62-bodies and contributes to inflammatory tolerance. (A) IP of SQSTM1 from MDMs treated with DHA for 16 h and IB analysis of KEAP1 and TAX1BP1; INPUT (10%). Data are representative of 3 independent experiments. (B) Immunostaining for SQSTM1 and KEAP1 in MDMs treated with DHA for 24 h (scale bars: 10 µm). (C) IB analysis and quantification (below) of SQSTM1 and KEAP1 in soluble and insoluble fractions from RAW264.7 cells after 16 h DHA, n = 4 (repeated measures ANOVA). (D) QRT-PCR analysis of CXCL10 or (E) CXCL11 in KEAP1 siRNA-transfected MDMs treated with DHA for 16 h and LPS for 3 h, n = 5 (Friedman test with Dunn's). (right) Comparison of the downregulating effect of DHA in control or KEAP1 siRNA-treated MDMs from (D) or (E) respectively, n = 5 (Mann Whitney test).
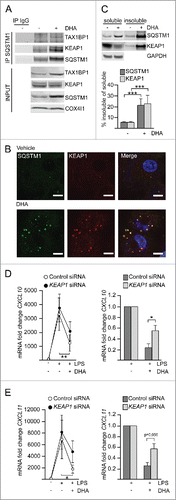
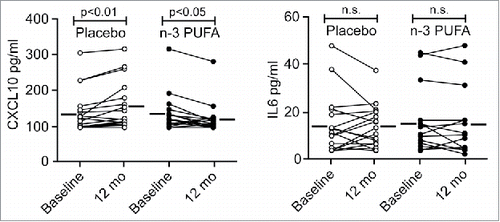
Figure 6. The plasma level of CXCL10 is reduced by n-3 PUFA supplementation but increased in the placebo group in hypertensive heart transplant recipients. The concentration of CXCL10 (left) and IL6 (right) was determined by ELISA in plasma of hypertensive heart transplant recipients isolated at baseline and after 12 mo of daily supplementation of placebo (corn oil, n = 19) or n-3 PUFAs (n = 16). Horizontal lines represent mean plasma levels. p < 0.01, p < 0.05 and nonsignificant (n.s.) were all vs baseline.