Figures & data
Figure 1. 3BDO could suppress the increase in CA7-4 expression caused by high glucose. (a) The scatter plot of microarray analysis depicts genes upregulated (red) and downregulated (green). CA7-4 level was decreased obviously, as indicated by the arrow. (b) Changes in CA7-4 level with treatment. (c) Information on CA7-4 in the human genome is available at https://lncipedia.org/db/transcript/lnc-CA7-4:1. (d,e) VECs were treated with 5.5 mM glucose (NG), 30 mM glucose (HG), and HG+3BDO (15, 30, 60 μM) for 24 h and 48 h. (f) VECs were treated with NG, 3BDO (0, 15, 30 μM) for 24 h. The level of CA7-4 was detected by qPCR. (*, p < 0.05; **, p < 0.01; n = 3.).
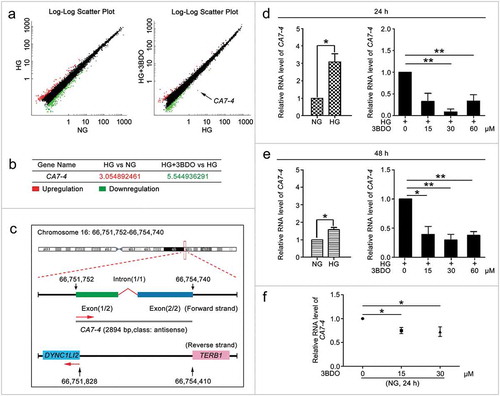
Figure 2. CA7-4 encouraged VEC autophagy. (a) VECs were transfected with empty vector pcDNA3.1 and CA7-4 (0.1, 0.25, 0.5 μg/ml) for 48 h. qPCR analysis of CA7-4 overexpression efficiency. (b) VECs were transfected with NC or siRNA-CA7-4-a, siRNA-CA7-4-b, and siRNA-CA7-4-c for 48 h. qPCR analysis of the RNA level of CA7-4. (c) After transfection with pcDNA3.1 or CA7-4 (0.25 μg/ml) for 24 h, VECs were treated with HG for 48 h. The distribution of LC3B in VECs was detected by immunofluorescence, and the proportion of cells containing > 5 LC3B puncta was investigated. Scale bar: 50 μm. Rescue experiment: VECs were transfected with pcDNA3.1 or CA7-4, then co-transfected with CA7-4 (0.1 μg/ml) and NC; CA7-4 (0.1 μg/ml) and si-CA7-4 (60 nM) for 24 h (d) or 48 h (e), then treated with HG (d) for 48 h. (f) VECs were treated with 3BDO (15 μM) for 48 h after transfection with CA7-4 (0.1 μg/ml) overnight. Western blot analysis of LC3B-II level. (*, p < 0.05; **, p < 0.01; n = 3.).
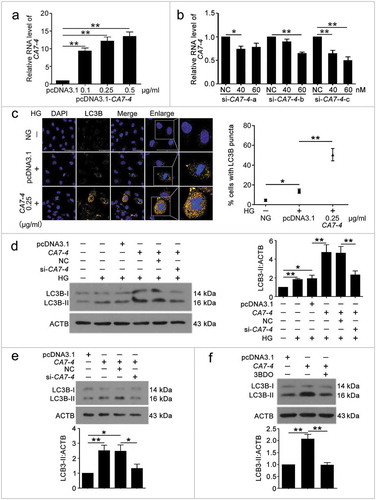
Figure 3. Overexpressing CA7-4 increased VEC apoptosis. (a–d) After being transfected with pcDNA3.1 or CA7-4; NC or si-CA7-4 overnight, VECs were treated with HG for 48 h, and the protein levels of BCL2, BAX and cleaved-PARP1 were detected by western blot. (e) VECs were treated with HG for 48 h after overexpression of CA7-4. Hoechst 33258 staining analysis of cell apoptosis. Scale bar: 30 μm. (f) VECs were transfected with pcDNA3.1 or CA7-4; co-transfected with CA7-4 (0.1 μg/ml) and NC or si-CA7-4 (60 nM) for 48 h. The level of cleaved-CASP3 was detected by western blot. (*, p < 0.05; **, p < 0.01; n = 3.).
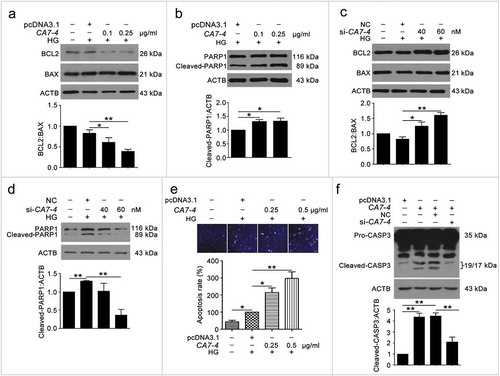
Figure 4. CA7-4 directly bonded with MIR877-3P and MIR5680. (a–d) qPCR analysis of MIR877-3P and MIR5680 in VECs after overexpression or knockdown CA7-4 for 48 h. (e–h) VECs were treated with NG, HG, HG+3BDO, or 3BDO (NG) for 24 h, qPCR analysis of MIR877-3P and MIR5680. (i,j) Luc-CA7-WT, Luc-CA7-4-MIR877-3P-Mut, and Luc-CA7-4- MIR5680-Mut plasmids were separately co-transfected into HEK293T cells with NC, MIR877-3P mimics or MIR5680 mimics for 24 h. Detecting luciferase activity. (k–m) RNA binding protein immunoprecipitation experiments were performed by using AGO2 antibody in VECs after treatment with NG, 3BDO (30 μM), HG and HG+3BDO (30 μM) for 24 h. IgG was used as a negative control. The fold enrichment of pulled-down CA7-4 was detected by qPCR analysis. (*, p < 0.05; **, p < 0.01; n = 3.).
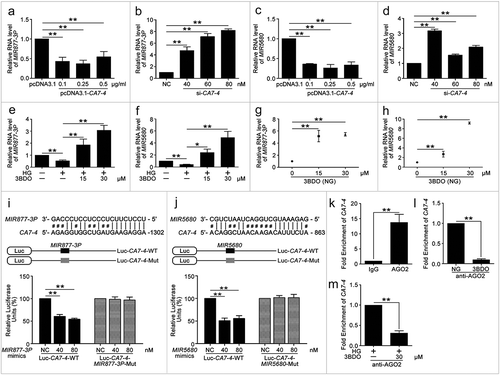
Figure 5. MIR877-3P inhibited VEC autophagy and apoptosis. After transfection with MIR877-3P mimics or inhibitor overnight, VECs were treated with HG for 48 h. (a,b) Western blot analysis of LC3B-II level. (c) TUNEL staining analysis of cell apoptosis. Scale bar: 100 μm. (d,e) Cell apoptosis was detected by Hoechst 33258 staining. Scale bar: 30 μm. (*, p < 0.05; **, p < 0.01; n = 3.).
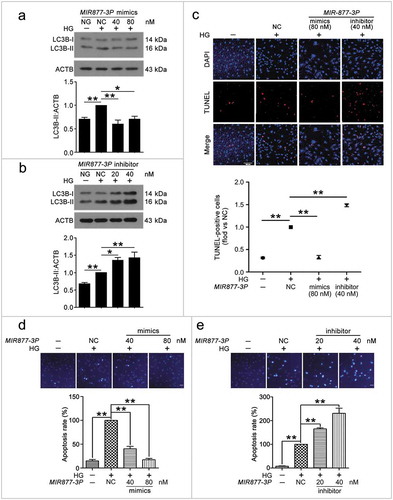
Figure 6. MIR5680 prevented VEC autophagy and apoptosis. VECs were treated with HG for 48 h after transfection with MIR5680 mimics or inhibitor overnight. (a) Western blot analysis of LC3B-II level. (b,c) Immunofluorescence analysis of the distribution of LC3B in VECs, and the proportion of cells containing > 5 LC3B puncta was analyzed. Scale bar: 20 μm. (d) TUNEL staining analysis of cell apoptosis. Scale bar: 50 μm. (*, p < 0.05; **, p < 0.01; n = 3.).
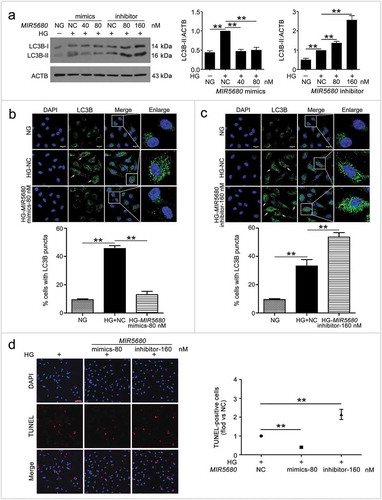
Figure 7. MIR877-3P targeted and inhibited CTNNBIP1. (a) Potential bind sites of MIR877-3P in CTNNBIP1 3ʹ UTR. (b) Luc-CTNNBIP1 3ʹ UTR-WT or Luc-CTNNBIP1 3ʹ UTR-Mut plasmid was co-transfected with NC or MIR877-3P mimics into HEK293T cells for 24 h. Luciferase activity was determined. (c) After transfection with NC, MIR877-3P inhibitor or mimics overnight, VECs were treated with HG for 48 h. Western blot analysis of CTNNBIP1. (d) VECs were treated with HG for 48 h after transfection with MIR877-3P mimics or co-transfection with MIR877-3P mimics and CA7-4. qPCR analysis of CTNNBIP1 mRNA level. (e,f) VECs were transfected with NC, MIR877-3P mimics or inhibitor for 48 h. (g) VECs were transfected with pcDNA3.1 or CA7-4 (0.1 μg/ml); co-transfected with CA7-4 (0.1 μg/ml) and NC or si-CA7-4 (60 nM) for 48 h. Western blot analysis of CTNNBIP1. (*, p < 0.05; **, p < 0.01; n = 3.).
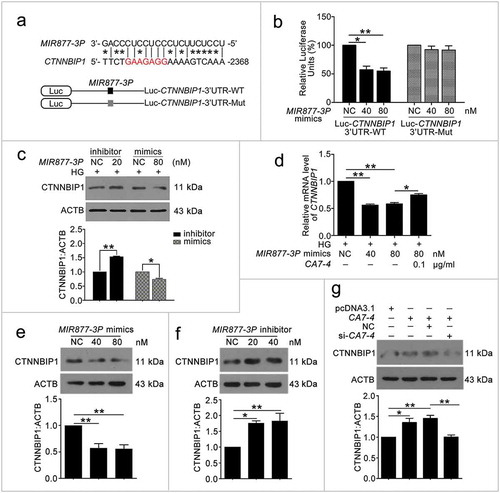
Figure 8. MIR5680 targeted DPP4 and decreased the mRNA level of it. (a) Possible binding sites of MIR5680 in DPP4 3ʹ UTR. (b) Luc-DPP4 3ʹ UTR-WT or Luc-DPP4 3ʹ UTR-Mut plasmids were co-transfected with NC or MIR5680 mimics into HEK293T cells for 24 h. Luciferase activity was determined. (c) After transfection with NC, MIR5680 mimics for 48 h, detecting the mRNA level of DPP4 by qPCR analysis. (d) VECs were treated with HG for 48 h after overexpression or knockdown of CA7-4. qPCR analysis of DPP4 mRNA level. (*, p < 0.05; **, p < 0.01; n = 3.).
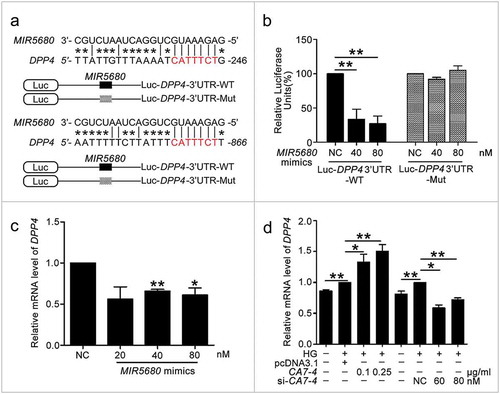
Figure 9. The protein level of DPP4 was negatively regulated by MIR5680. (a,b) VECs were transfected with MIR5680 mimics or inhibitor for 48 h. (c) VECs were transfected with pcDNA3.1 or CA7-4 (0.1 μg/ml); co-transfected with CA7-4 (0.1 μg/ml) and NC or si-CA7-4 (60 nM) for 48 h. (d,e) After transfection with NC, MIR5680 mimics or inhibitor overnight, VECs were treated with HG for 48 h. Western blot analysis of DPP4 level. (*, p < 0.05; **, p < 0.01; n = 3.).
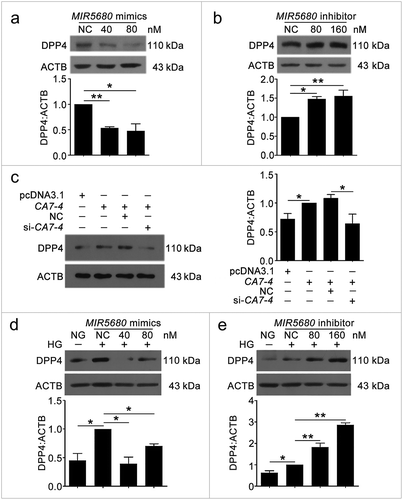
Figure 10. The knockdown of CTNNBIP1 and DPP4 reduced VEC autophagy and apoptosis caused by high glucose. (a–d) After transfection with NC, si-CTNNBIP1 or si-DPP4 overnight, VECs were treated with high glucose for 48 h. The protein levels of CTNNBIP1, CTNNB1, cleaved-CASP3, LC3B-II, DPP4, p-AMPK were analyzed by western blot. (*, p < 0.05; **, p < 0.01; n = 3.).
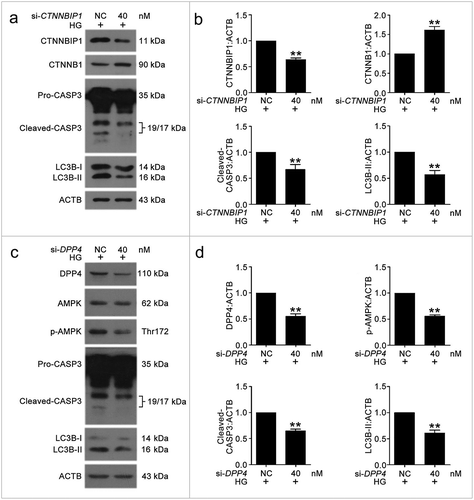
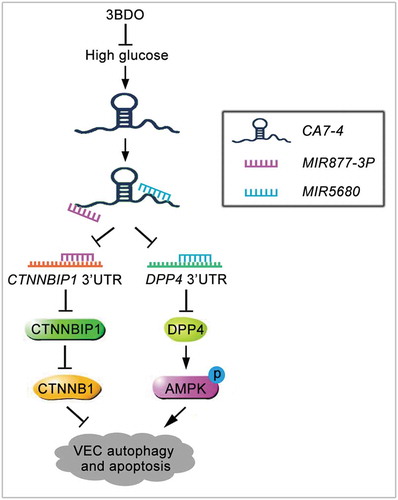
Figure 11. Schematic of CA7-4 regulating VEC autophagy and apoptosis caused by high glucose. High glucose induces VEC autophagy and apoptosis. In this progress, upregulated CA7-4 as a miRNA sponge to decoy MIR877-3P and MIR5680, promote the expression of CTNNBIP1 and DPP4, decrease the level of CTNNB1 and increase AMPK phosphorylation. 3BDO can attenuate high glucose-induced VEC autophagy and apoptosis by weakening the upregulation of CA7-4.