Figures & data
Figure 1. Atg30-independent degradation of PTS receptors. (A, B) Pex5, Pex7, and Pex20 were degraded in both (A) WT and (B) atg30∆ cells. Peroxisome proliferation were induced by growing cells in oleate medium, followed by adaptation in glucose medium without nitrogen (SD-N) for the indicated times. Crude cell lysates were extracted by TCA precipitation, resolved by SDS-PAGE, and detected with anti-Pot1, anti-Pex5, and anti-HA (for Pex7 and Pex20). (C) Quantification of the degradation of Pex5, Pex7, and Pex20 (from A and B) using ImageJ and expressed as the percentage of the total signal at 0 h. The results represent the mean and standard deviation (SD) of triplicate biological replicates.
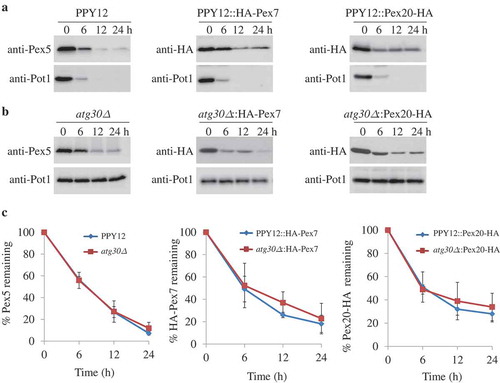
Figure 2. Pex5 and Pex7 are removed by selective autophagy. (A, B) Peroxisomes were induced by growing atg30∆ cells expressing Pex5K22R, Pex5K22R and UbK48R, Pex20K19R, or Pex20K19R and UbK48R in oleate medium. Subsequently, cells were starved for pexophagy experiments. Samples were taken at the indicated time points under starvation conditions and analyzed by immunoblots. (C, D) The degradation of Pex5 (from A) and Pex20 (from B) from triplicate biological repeats was quantified in ImageJ and expressed as the percentage of total signal at time 0 h. (E, F) Oleate-grown WT, SMD1163, atg1∆, atg8∆, atg11∆, atg17∆, atg11∆ atg17∆ cells or strains expressing HA-Pex7 were adapted to glucose medium without nitrogen (SD-N). Samples were collected at the indicated time points under starvation conditions for immunoblotting analysis. (G, H) The degradation of Pex5 (from E) and Pex7 (from F) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of total signal at 0 h.
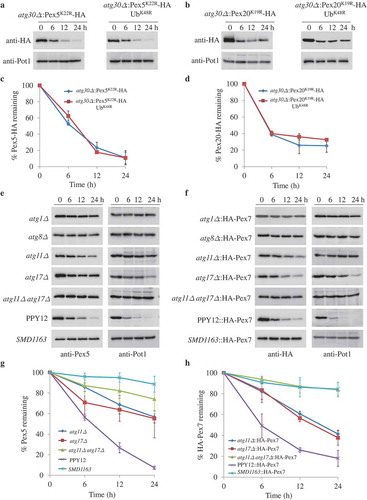
Figure 3. The absence of Pex5 or Pex7 under pexophagy conditions is irrelevant for the turnover of other PTS receptors. (A) Upon starvation, Pex5 was degraded in pex7∆, pex20∆, and pex7∆ pex20∆ cells. (B) Pex7 was degraded in pex5∆, pex20∆, and pex5∆ pex20∆ cells under pexophagy conditions. Pex2, a PMP, was used as a positive control for the degradation of peroxisomes. The protein levels of Pex2, Pex5, and HA-Pex7 were analyzed by immunoblotting with anti-Pex2, anti-Pex5, and anti-HA (for Pex7). (C, D) The degradation of Pex5 (from A) and Pex7 (from B) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of total signal at 0 h.
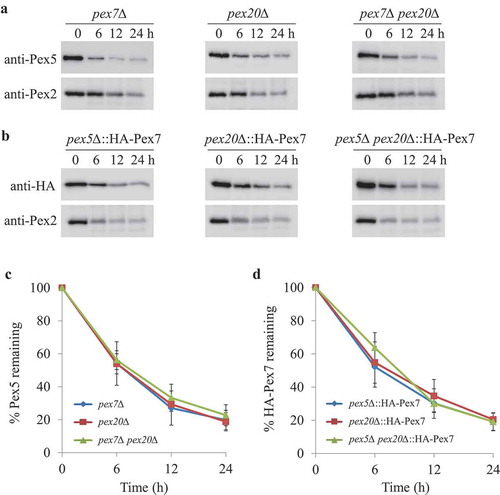
Figure 4. Degradation of Pex7 depends on Pot1, which is selectively degraded when mislocalized. (A) Peroxisomes were induced by growing pot1∆ expressing HA-Pex7 and pex7∆ expressing HA-Pex7A248R in oleate medium. Subsequently, cells were transferred to starvation medium for pexophagy experiments. Samples were taken at the indicated time intervals under starvation condtions and analyzed by immunoblotting with anti-HA, anti-Pot1, and anti-Pex2 antibodies. (B) The degradation of Pex7 (from A) from triplicate biological repeats was quantified in ImageJ and expressed as the percentage of total signal at time 0 h. (C) Fluorescence analysis of WT, pex7∆, pex7∆ atg11∆, and pex7∆ atg11∆ atg17∆ strains expressing Pot1-GFP under starvation conditions in the presence of the vacuolar stain, FM 4–64. The differential interference contrast (DIC) and GFP images are shown. (D) Strains shown in (C) were used for GFP cleavage assays. Samples were analyzed by immunoblotting with anti-GFP. (E) The full-length Pot1-GFP and the cleaved GFP under starvation conditions (from D) were quantified using tripliciate biological repeats and ImageJ and expressed as the ratio between cleaved GFP and total GFP signals at each time point. *: non-specific band.
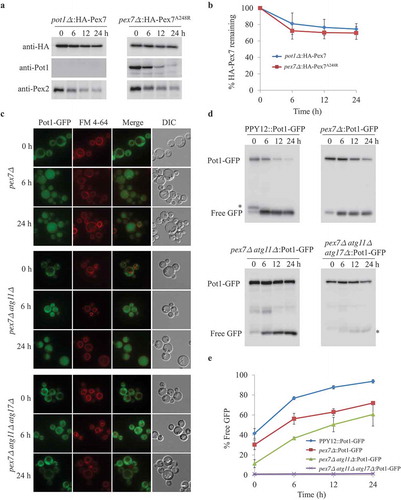
Figure 5. Pex5 is degraded together with its cargoes. (A) Peroxisomes were induced by growing WT, SMD1163, aox1∆ aox2∆, fox2∆, aox1∆ aox2∆ fox2∆ or pex5∆ and aox1∆ aox2∆ pex5∆, each expressing the Pex5N460K mutant in oleate medium. Subsequently, cells were transferred to starvation medium for pexophagy experiments. Samples were taken at the indicated time points under starvation conditions and analyzed by immunoblotting with anti-Pex5 and anti-Pot1. (B) The degradation of Pex5 (from A) from triplicate biological repeats was quantified using ImageJ and expressed as the percent of the total signal at 0 h.
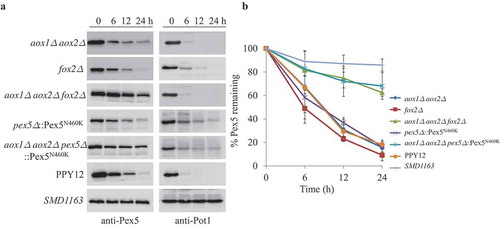
Figure 6. Cytosolic Fox2 and Pmp20 are degraded by selective autophagy. (A, B) Oleate-grown pex5∆, pex5∆ atg11∆, and pex5∆ atg11∆ atg17∆ expressing GFP-Fox2 (A) or GFP-Pmp20 (B) from their endogenous promoters were transferred to starvation medium for pexophagy experiments in the presence of the vacuolar stain, FM 4–64. Fluorescence pictures were taken at 0, 6, and 24 h after starvation. (C) WT, pex5∆, pex5∆ atg11∆, and pex5∆ atg11∆ atg17∆ expressing GFP-Pmp20 were used for GFP cleavage assays. Samples were analyzed by immunoblotting with anti-GFP. A slightly longer exposure time for the pex5Δ atg11∆::GFP-Pmp20 blot to see the free GFP band. (D) The full-length GFP-Pmp20 and free GFP under starvation conditions (from C) were quantified using tripliciate biological repeats and ImageJ and expressed as the ratio between cleaved GFP and total GFP signals at each time point. *: non-specific band.
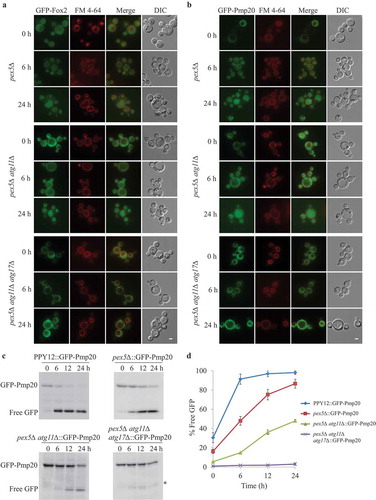
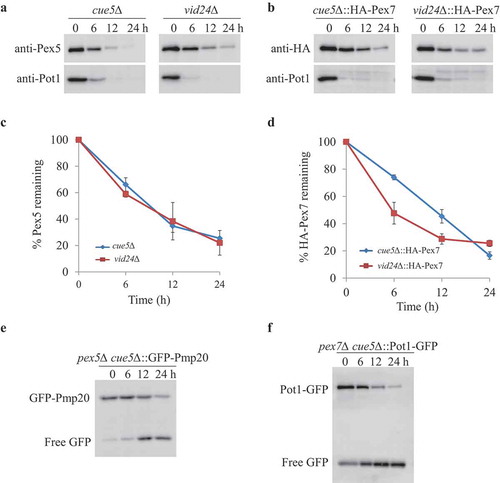
Figure 7. Aggrephagy and Vid pathways were not involved in removing Pex5, Pex7 and cytosolic peroxisomal cargoes. (A, B) oleate-grown cue5Δ and vid24∆ (A) or cue5∆ and vid24∆ cells expressing HA-Pex7 from its native promoter (B) were shifted to starvation medium for pexophagy experiments. (C, D) The degradation of Pex5 and Pex7 (from A and B) was quantified in ImageJ and expressed as the percentage of the total signal at time 0 h. The results represent the mean and standard deviation (SD) of triplicate biological repeats. (E, F) Oleate-grown pex5∆ cue5∆ cells expressing GFP-Pex20 (E) and pex7∆ cue5∆ cells expressing Pot1-GFP (F) were transferred to stavation medium for GFP cleavage assays. Samples were taken at the indicated time points under starvation condtions and analyzed by immunoblotting with anti-GFP.