Figures & data
Figure 1. DIPK2A is localized to the lysosomes and late endosomes. (A) Sucrose gradient separation analysis of DIPK2A in HeLa cells by western blotting (WB). (B) HeLa cells were co-transfected with DIPK2A-mCherry (red) and GFP-RAB7A (late endosome marker, green), GFP-LAMP1 (lysosome marker, green) or GFP-RAB5A (early endosome marker, green). The localization of the DIPK2A (red) with endosomes (green) or lysosomes (green) was analyzed by fluorescence microscopy. The boxed areas are magnified. Colocalized puncta are marked by arrowheads. Scale bar: 10 μm. (C) HeLa cells were transfected with DIPK2A-mCherry (red) and immunostained for LAMP1 (green). The localization of DIPK2A (red) and lysosomes (green) was analyzed by super-resolution fluorescence microscopy, structure illumination microscopy (SIM). The boxed areas are magnified. Scale bar: 2 μm. (D) DIPK2A-mCherry knock-in HEK293T cells were transfected with GFP-LAMP1, and the localization of DIPK2A (red) and LAMP1 (green) was analyzed by fluorescence microscopy. The boxed areas are magnified. Colocalized puncta are marked by arrowheads. Scale bar: 10 μm. See also Figure S1.
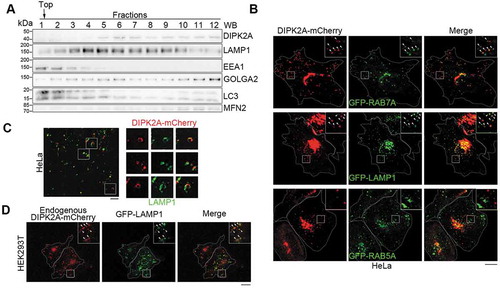
Figure 2. DIPK2A promotes autophagy and mitophagy. (A-C) Wild-type (WT) or DIPK2A knockout (KO) HeLa cells were cultured in normal medium, EBSS, or EBSS plus CQ for 2 h, harvested, and subjected to western blotting (WB) analysis. ACTB was used as a loading control (A). The relative band intensity of LC3-II (B) and SQSTM1 (C) was compared with the loading control based on 3 independent experiments. (D and E) Wild-type (WT) or DIPK2A knockout (KO) HeLa cells transfected with GFP-PRKN were treated with DMSO or 10 μM CCCP for 24 h, harvested, and subjected to western blotting (WB) analysis. ACTB was used as a loading control (D). The relative protein levels were compared with the loading control based on 3 independent experiments (E). (F-H) HeLa cells co-transfected with mRFP-GFP-LC3 and DIPK2A-BFP (blue) or the control vector were cultured under normal or EBSS (2 h) conditions and imaged by fluorescence microscopy (F). Scale bar: 10 μm. The percentage of GFP+ mRFP+ puncta (G), and the numbers of GFP+ mRFP+ and GFP– mRFP+ puncta (H) were determined (n ≥ 75 cells). (I-K) Wild-type (WT) or DIPK2A knockout (KO) HeLa cells were transfected with mRFP-GFP-LC3, cultured under normal or EBSS (2 h) conditions, and imaged by fluorescence microscopy (I). Scale bar: 10 μm. The percentage of GFP+ mRFP+ puncta (J), and the numbers of GFP+ mRFP+ and GFP– mRFP+ puncta (K) were determined (n ≥ 40 cells). For (G, H, J and K), the results are expressed as mean ± S.D. Statistical significance (p-value) was determined by one-way ANOVA (listed at the top). See also Figure S2.
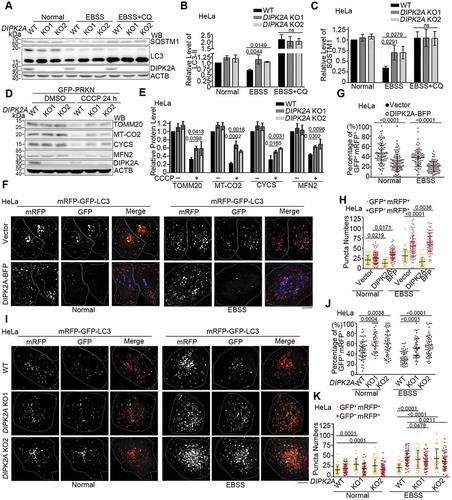
Figure 3. VAMP7B interacts with DIPK2A and STX17. (A and B) HEK293T cells transfected with the indicated vectors were immunoprecipitated (IP) by the indicated antibodies and subjected to western blotting (WB) analysis. (C) HEK293T cells transfected with DIPK2A-HA and the indicated VAMP7B mutants were immunoprecipitated (IP) by anti-FLAG antibody and subjected to western blotting (WB) analysis. FL, full length; numbers indicate amino acids. (D) HEK293T cells co-transfected with MYC-STX17 and the indicated vectors were immunoprecipitated (IP) by anti-FLAG antibody and subjected to western blotting (WB) analysis. (E and F) HEK293T cells co-transfected with V5-STX17 and FLAG-tagged VAMP7 truncates (E) or VAMP7A deletion mutants (F) were immunoprecipitated (IP) by anti-FLAG antibody and subjected to western blotting (WB) analysis. FL, full length; numbers indicate amino acids. (G and H) HEK293T cells co-transfected with MYC-tagged STX17 mutants and 3× FLAG-VAMP7A (G) or 3× FLAG-VAMP7B (H) were immunoprecipitated (IP) with anti-MYC antibody and subjected to western blotting (WB) analysis. FL, full length; numbers indicate amino acids. (I) Diagram of the interaction among DIPK2A, STX17, VAMP7A and VAMP7B. See also Figure S3.
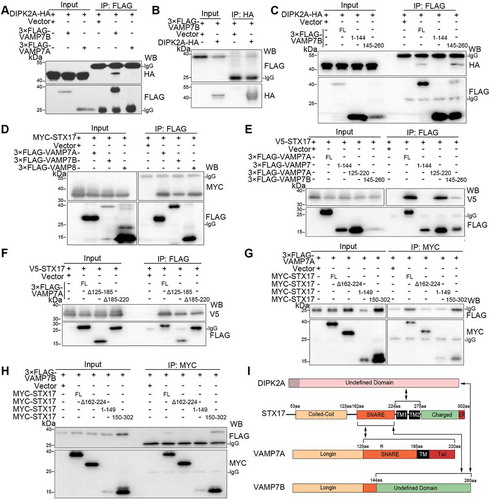
Figure 4. VAMP7B inhibits autophagosome-lysosome fusion by ameliorating interaction between VAMP7A and STX17. (A) HeLa cells transfected with GFP-LC3 (green), 3× FLAG-VAMP7B and V5-SXT17 were treated with EBSS in the presence of bafilomycin A1 (Baf. A1) for 2 h before probed with anti-FLAG and anti-V5 antibodies, and subjected to PLA (red). Samples were observed by structure illumination microscopy (SIM). The boxed areas are magnified. Scale bar: 2 μm. (B-D) VAMP7 knockout (KO) HeLa cells co-transfected with mRFP-GFP-LC3 and the indicated vectors were cultured under normal or EBSS (2 h) conditions and imaged by fluorescence microscopy (B). Scale bar: 10 μm. The percentage of GFP+ mRFP+ puncta (C) and the numbers of GFP+ mRFP+ and GFP– mRFP+ puncta (D) were determined (n ≥ 50 cells). (E) HEK293T cells transfected with the indicated vectors were immunoprecipitated (IP) using an anti-MYC antibody and subjected to western blotting (WB) analysis. The relative band intensities are marked below. For (C and D), the results are expressed as mean ± S.D. The statistical significance (p-value) was determined by one-way ANOVA (listed at the top). See also Figure S4.
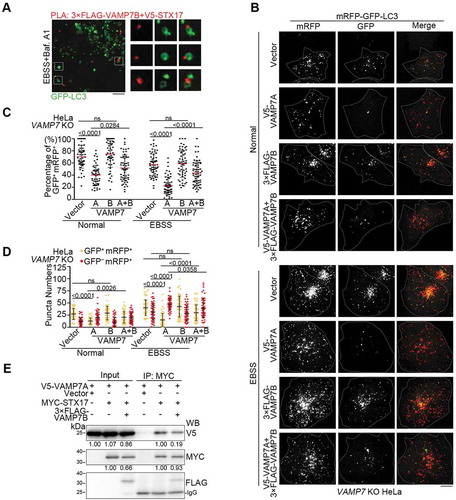
Figure 5. DIPK2A attenuates the interaction between STX17 and VAMP7B while enhancing the interaction between SXT17 and VAMP7A. (A) HEK293T cells transfected with the indicated vectors were immunoprecipitated (IP) using anti-MYC and anti-FLAG antibodies and subjected to western blotting (WB) analysis. The relative band intensities are marked below. (B and C) HeLa cells were transfected with 3× FLAG-VAMP7B and V5-SXT17 in the presence of DIPK2A-BFP (blue) or not. PLA (red) was performed by probing with anti-FLAG and anti-V5 antibodies (B). Scale bar: 10 μm. The number of PLA-positive puncta were quantified (n ≥ 50 cells) (C). (D) HeLa cells transfected with DIPK2A-3× FLAG or the control vector for 36 h were immunoprecipitated by an anti-STX17 antibody or rabbit IgG, and subjected to western blotting (WB) analysis. The relative band intensities are marked below. (E-G) Wild-type (WT), STX17 knockout (KO) and VAMP7 knockout (KO) HeLa cells co-transfected with mRFP-GFP-LC3 and the indicated vectors were cultured under normal or EBSS (2 h) conditions and imaged by fluorescence microscopy (E). Scale bar: 10 μm. The percentage of GFP+ mRFP+ puncta (F), and the numbers of GFP+ mRFP+ and GFP– mRFP+ puncta (G) were determined (n ≥ 40 cells). For (C, F and G), the results are expressed as mean ± S.D. The statistical significance (p-value) was determined by unpaired two-tailed Student’s t-tests (C) or two-way ANOVA (F and G) (listed at the top). See also Figure S4.
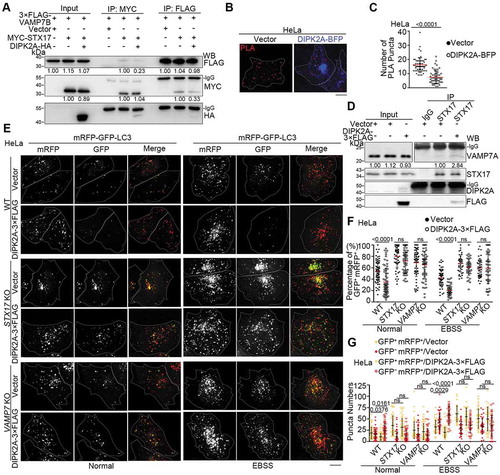
Figure 6. STX17 interacts with DIPK2A via two transmembrane domains. (A and B) HEK293T cells co-transfected with 3× FLAG-STX17 and DIPK2A-HA were immunoprecipitated (IP) by the indicated antibodies and subjected to western blotting (WB) analysis. (C) HeLa cells were collected and lysed. Endogenous proteins were immunoprecipitated (IP) by an anti-DIPK2A antibody or rabbit IgG (as a control). (D-F) HEK293T cells co-transfected with DIPK2A-3× FLAG and MYC-tagged STX17 truncates were immunoprecipitated (IP) by an anti-MYC antibody and subjected to western blotting (WB) analysis. FL, full length; numbers indicate amino acids. (G) HEK293T cells transfected with the indicated vectors were immunoprecipitated (IP) by an anti-FLAG antibody and subjected to western blotting (WB) analysis. FL, full length; numbers indicate amino acids. The relative band intensities are marked below.
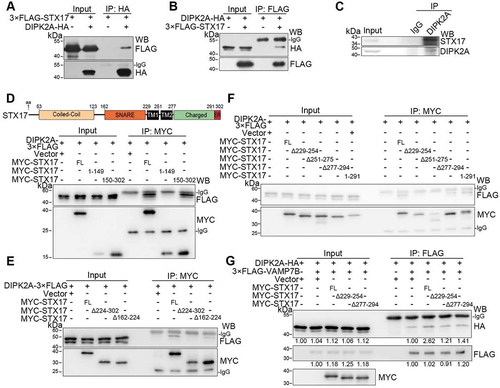
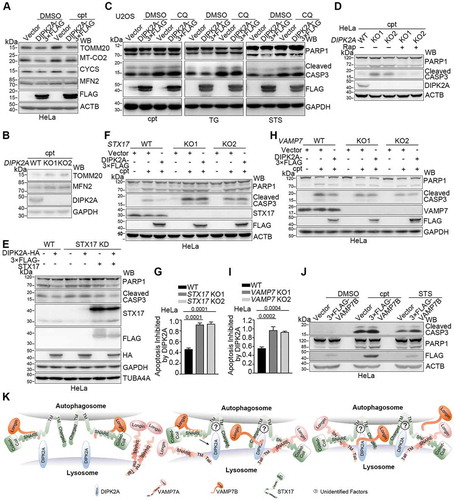
Figure 7. DIPK2A inhibits apoptosis. (A) U2OS cells were seeded in 96-well cell plates and transfected with the control vector or DIPK2A-3× FLAG. 1 μM cpt was added 24 h after transfection. The cells were analyzed by MTT assay. (B) HeLa cells were transfected with the control vector or DIPK2A-3× FLAG and treated with 500 ng/mL TG, 2 mM H2O2, 10 nM STS, or 1 μM cpt for 24 h. Cell lysates were subjected to western blotting (WB) analysis using the indicated antibodies. GAPDH was used as a loading control. (C) Wild-type (WT) or DIPK2A knockout (KO) U2OS cells were seeded in 96-well cell plates and treated with 1 μM cpt. The cells were analyzed by MTT assay. (D) Wild-type (WT) and DIPK2A knockout (KO) U2OS cells were treated with 500 ng/mL TG, 2 mM H2O2, 10 nM STS, or 1 μM cpt for 24 h and subjected to western blotting (WB) analysis. GAPDH was used as a loading control. (E) Wild-type (WT) or DIPK2A knockout (KO) U2OS cells were transfected with the indicated vectors and treated with 500 ng/mL TG, 2 mM H2O2, 10 nM STS or 1 μM cpt for 24 h. Cell lysates were subjected to western blotting (WB) analysis. GAPDH was used as a loading control.
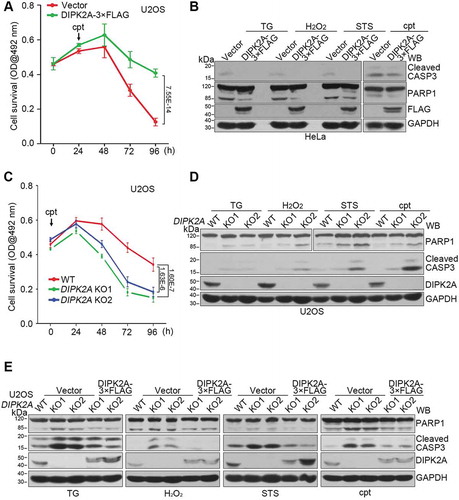
Figure 8. Inhibition of apoptosis by DIPK2A depends on its ability to regulate autophagy and on STX17-VAMP7. (A) HeLa cells transfected with the control vector or DIPK2A-3× FLAG were treated with 1 μM cpt for 24 h. The cell lysates were subjected to western blotting (WB) analysis. ACTB was used as a loading control. (B) Wild-type (WT) and DIPK2A knockout (KO) cells were treated with 1 μM cpt for 24 h and subjected to western blotting (WB) analysis. GAPDH was used as a loading control. (C) HeLa cells transfected with the control vector or DIPK2A-3× FLAG were treated with 1 μM cpt, 500 ng/mL TG or 10 nM STS. DMSO or CQ was added at the same time. The cell lysates were subjected to western blotting (WB) analysis. GAPDH was used as a loading control. (D) Wild-type or DIPK2A knockout (KO) HeLa cells transfected with the control vector or DIPK2A-3× FLAG were treated with 1 μM cpt, and with or without 20 nM rapamycin (Rap) for 12 h. The cell lysates were subjected to western blotting (WB) analysis. ACTB was used as a loading control. (E) Wild-type (WT) or STX17-constitutively-knockdown (STX17 KD) HeLa cells were transfected with the indicated vectors and treated with 1 μM cpt for 24 h. The cell lysates were subjected to western blotting (WB) analysis. TUBA4A was used as a loading control. (F and G) Wild-type (WT) or STX17 knockout (KO) HeLa cells were transfected with the indicated vectors, and treated with or without 1 μM cpt for 24 h. The cell lysates were subjected to western blotting (WB) analysis. ACTB was used as a loading control (F). Inhibition of the relative apoptosis level (cleaved PARP1) by DIPK2A was determined based on 3 independent experiments and normalized based on the control groups (without cpt treatment) (G). (H and I) Wild-type (WT) or VAMP7 knockout (KO) HeLa cells were transfected with the indicated vectors, and treated with or without 1 μM cpt for 24 h. The cell lysates were subjected to western blotting (WB) analysis using the indicated antibodies. GAPDH was used as a loading control (H). Inhibition of the relative apoptosis level (cleaved PARP1) by DIPK2A was determined based on 3 independent experiments and normalized based on the control groups (without cpt treatment) (I). (J) HeLa cells transfected with the control vector or 3× FLAG-VAMP7B were treated with DMSO, 1 μM cpt or 10 nM STS for 24 h, after which they were subjected to western blotting (WB) analysis. ACTB was used as a loading control. (K) Model of STX17-, VAMP7- and DIPK2A-mediated autophagosome-lysosome fusion. VAMP7B binds to the SNARE domain of STX17 through its C terminus and inhibits binding of VAMP7A. Once DIPK2A binds to the TM domain of STX17, VAMP7B detaches from the SNARE domain of STX17, while VAMP7A binds to this domain. The simultaneous binding of VAMP7B and DIPK2A to different domains of STX17 enhances the binding between VAMP7B and DIPK2A. Some unidentified factors may also participate in this process.