Figures & data
Figure 1. Knockdown of Mir26a decreases autophagy to impair heart function. (A) Downregulation of Mir26a in the peri-infarct area determined by qRT-PCR. n = 6. *p < 0.05 vs. Sham. (B) Downregulation of Mir26a in cultured neonatal mouse cardiomyocytes (NMCMs) treated with 200 μmol/L H2O2 for 12 h. n = 5. **p < 0.01 vs. Control. (C) Knockdown of Mir26a inhibited autophagic flux in NMCMs. NMCMs were transfected with AMO-26a for 24 h, followed by transfection of tandem-LC3 construct (GFP-mRFP-LC3) for another 24 h. The images were obtained using a confocal microscope. The yellow and red puncta represent autophagosomes and autolysosomes, respectively. The data were obtained from 3 independent experiments and 10 cells were scored in each experiment. *p < 0.05. NC, negative control of AMO-26a. (D) Knockdown of Mir26a increased the level of SQSTM1/p62 protein and reduced the expression of LC3-II, as determined by western blot. n = 5. *p < 0.05 vs. Control. NC, negative control of AMO-26a. (E) Downregulation of Mir26a in Mir26a KD mice. n = 6. **p < 0.01 vs. WT. (F) Knockdown of Mir26a impaired heart function in mice. n = 15. **p < 0.01 vs. WT. (G) Knockdown of Mir26a resulted in the deregulation of autophagy-related proteins in mouse hearts. n = 6. *p < 0.05 vs. WT.
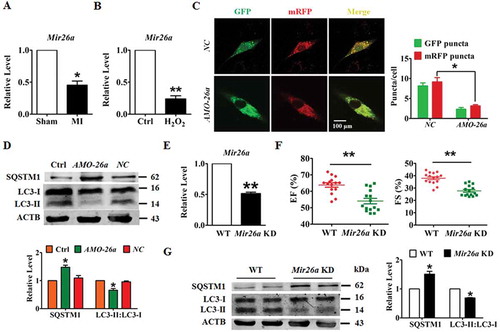
Figure 2. Forced expression of Mir26a protects cardiomyocytes from injury by activating autophagy. (A) Overexpression of Mir26a alleviated H2O2-induced inhibition of cell viability in NMCMs. This effect was reversed by application of 3-MA, an autophagic antagonist. n = 8. **p < 0.01 vs. Control; ##p < 0.01 vs. H2O2; &&p < 0.01 vs. H2O2+ Mir26a. MirNC, negative control of Mir26a. (B) Forced expression of Mir26a restored the inhibition of autophagy induced by H2O2. Data were obtained from 3 independent experiments and 10 cells were scored in each experiment. **p < 0.01. MirNC, negative control of Mir26a. (C) Overexpression of Mir26a improved heart function (n = 9. *p < 0.05 vs. Sham; #p < 0.05 vs. MI) and reduced the infarct size (both IA/NIA and IA/LV) in MI mice (D) (n = 9. **p < 0.01 vs. MI+ago MirNC). (E) Overexpression of Mir26a enhanced autophagic activity in MI mice. n = 5. **p < 0.01 vs. MI. AgomiR-NC, negative control of agoMir26a. (F) Overexpression of Mir26a normalized the dysregulated expression of autophagy-related proteins in MI mice. n = 6. *p < 0.05 vs. Sham; #p < 0.05 vs. MI. Ago MirNC, negative control of agoMir26a.
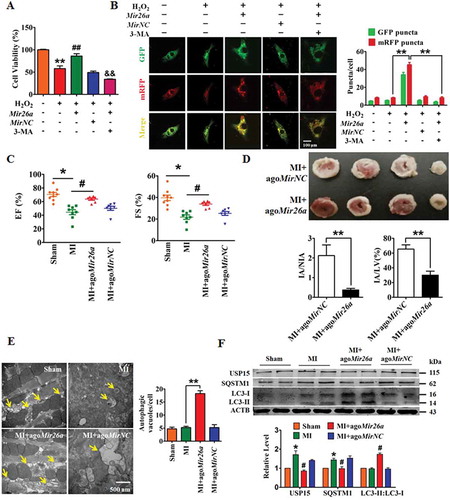
Figure 3. Usp15 is a direct target of Mir26a. (A) Sequence alignment showing complementarity between mouse, rat, and human Mir26a and Usp15 genes. The matched base pairs in the seed region are outlined by red and green. (B) Increased expression of USP15 in infarct border area in MI mice. n = 6. **p < 0.01 vs. Sham. (C) Increased expression of USP15 in NMCMs after treatment with 200 μmol/L H2O2 for 12 h. n = 6. (D) Luciferase reporter activity of chimeric vectors carrying a luciferase gene and a fragment of the Usp15 3′-UTR containing the wild-type or mutant binding sites for Mir26a were detected in HEK293 cell line. n = 6. *p < 0.05. MirNC, negative control of Mir26a. (E) Overexpression of Mir26a depressed protein levels of USP15 (n = 6. *p < 0.05 vs. Control. MirNC, negative control of Mir26a; NC, negative control of AMO-26a), (F) but did not affect its mRNA levels (n = 6). (G) Upregulation of USP15 in Mir26a KD mice, the three lanes of WT and KD correspond to three different mice. n = 6. **p < 0.01 vs. Sham. (H) Forced expression of Mir26a inhibited expression of SQSTM1 and promoted the transition of LC3-I into LC3-II. n = 6. *p < 0.05 vs. Control. MirNC, negative control of Mir26a.
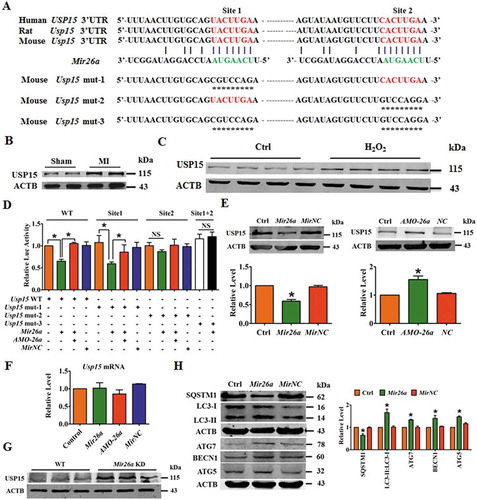
Figure 4. LncRNA 2810403D21Rik/Mirf regulates the expression and activity of Mir26a. (A) Upregulation of 2810403D21Rik/Mirf in MI mice. n = 6. **p < 0.01 vs. Sham. (B) Upregulation of 2810403D21Rik/Mirf in NMCMs after treatment with 200 μmol/L H2O2 for 12 h. n = 6. **p < 0.01 vs. Control. (C) In situ hybridization showed the location of 2810403D21Rik/Mirf in NMCMs. (D) Overexpression of 2810403D21Rik/Mirf downregulated Mir26a. n = 6. *p < 0.05 vs. Control. pcDNA3.1, negative control. (E) Silencing of 2810403D21Rik/Mirf by the specific shRNA upregulated Mir26a. The black bar indicates three different shRNA sequences against 2810403D21Rik/Mirf. n = 6. **p < 0.01 vs. sh-Scramble. sh-Scramble, negative control of sh-2810403D21Rik/Mirf. (F) 2810403D21Rik/Mirf contained a sequence domain complementary to the seed motif of Mir26a. (G and H) 2810403D21Rik/Mirf binds to Mir26a and regulates its activity in NMCMs. n = 6. *p < 0.05, **p < 0.01. (I) Luciferase reporter activities of chimeric vectors carrying luciferase gene and a fragment of 2810403D21Rik/Mirf RNA containing WT binding site or mutant binding site for Mir26a were detected in HEK293 cell line. n = 6. *p < 0.05. pcDNA3.1, negative control of 2810403D21Rik/Mirf. sh-Scramble, negative control of sh-2810403D21Rik/Mirf. MirNC, negative control of Mir26a. (J) RNA affinity isolation in NMCMs using biotin-labeled 2810403D21Rik/Mirf probes. Western blot was used to detected enrichments of AGO2. n = 3. (K) Mir26a was pulled down by 2810403D21Rik/Mirf probe, and the expression of Mir26a was analyzed by qRT-PCR. Cardiomyocytes were transfected with biotinylated Mir26a and then harvested for biotin-based affinity-isolation assay. n = 6. **p < 0.01. Scramble, negative control of 2810403D21Rik/Mirf. (L) 2810403D21Rik/Mirf was pulled down by Mir26a as analyzed by real-time RT-PCR. n = 6. **p < 0.01. MirNC, negative control of Mir26a. (M) The product of real-time RT-PCR was identified by agarose gel electrophoresis. MirNC, negative control of Mir26a.
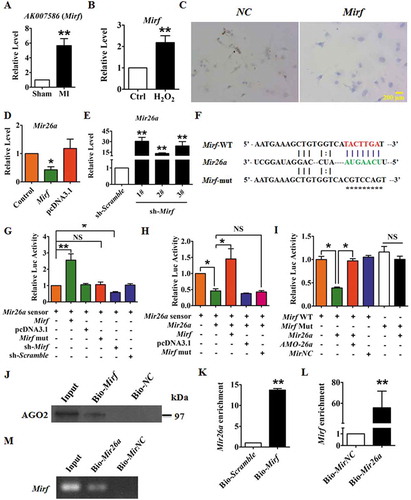
Figure 5. 2810403D21Rik/Mirf modulated autophagy by regulating Mir26a and Usp15, promoting cardiac injury in NMCMs. (A) Overexpression of 2810403D21Rik/Mirf decreased viability of NMCMs; this effect was attenuated by Mir26a or si-Usp15. n = 5. *p < 0.05. pcDNA3.1, negative control of 2810403D21Rik/Mirf. MirNC, negative control of Mir26a. si-Scramble, negative control of si-Usp15. (B) 2810403D21Rik/Mirf aggravated H2O2-induced cardiac injury in NMCMs. n = 6. *p < 0.05. pcDNA3.1, negative control of 2810403D21Rik/Mirf. MirNC, negative control of Mir26a. (C) Overexpression of 2810403D21Rik/Mirf blocked the autophagic activity of NMCMs. n = 10. *p < 0.05, **p < 0.01. pcDNA3.1, negative control of 2810403D21Rik/Mirf. MirNC, negative control of Mir26a. si-Scramble, negative control of si-Usp15. (D) Overexpression of 2810403D21Rik/Mirf produced anti-autophagic action through regulation of various relevant proteins, which was abolished by Mir26a. n = 5. *p < 0.05 vs. pcDNA3.1; #p < 0.05 vs. 2810403D21Rik/Mirf. pcDNA3.1, negative control of 2810403D21Rik/Mirf. MirNC, negative control of Mir26a.
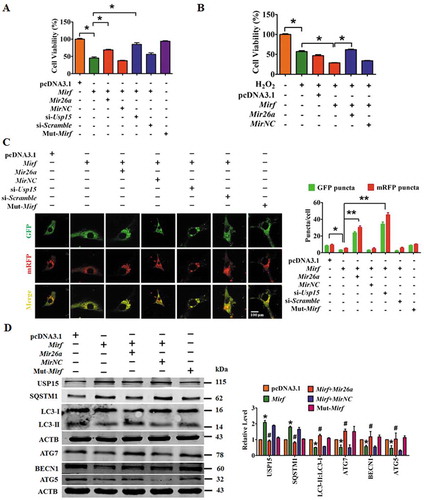
Figure 6. Inhibition of 2810403D21Rik/Mirf antagonizes H2O2-induced injury in NMCMs through modulation of autophagy by increasing Mir26a. (A) Silencing of 2810403D21Rik/Mirf alleviated H2O2-induced cardiac injury, which was reversed by knockdown of Mir26a by AMO-26a or inhibition of autophagy by 3-MA in NMCMs. n = 6. *p < 0.05. sh-Scramble, negative control of sh-2810403D21Rik/Mirf. NC, negative control of AMO-26a. (B) Inhibition of autophagy by si-Atg5, chloroquine or pepstatin A/E64d mitigated the pro-survival effects of 2810403D21Rik/Mirf silencing in H2O2-treated NMCMs. n = 6. *p < 0.05. (C) Knockdown of 2810403D21Rik/Mirf restored abnormal expression of autophagy-related proteins induced by H2O2. n = 5. *p < 0.05 vs. or Control; #p < 0.05 vs. H2O2; &p < 0.05 vs. H2O2+ sh-2810403D21Rik/Mirf. sh-Scramble, negative control of sh-2810403D21Rik/Mirf. NC, negative control of AMO-26a.(D) Silencing of 2810403D21Rik/Mirf promoted autophagy through increasing expression of Mir26a. The data were obtained from 3 independent experiments and 10 cells were scored in each experiment. **p < 0.01. sh-Scramble, negative control of sh-2810403D21Rik/Mirf. NC, negative control of AMO-26a.
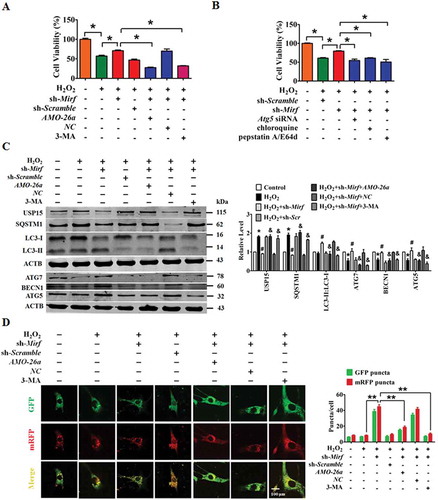
Figure 7. Overexpression of f-2810403D21Rik/Mirf provokes myocardial injury in cultured cardiomyocytes by inhibiting endogenous Mir26a. (A) Introduction of f-2810403D21Rik/Mirf, a 25 nt fragment of the 2810403D21Rik/Mirf sequence which contained the binding sites of Mir26a, inhibits the viability of NMCMs. n = 6. *p < 0.05 vs. Scramble; #p < 0.05 vs. f-2810403D21Rik/Mirf. Scramble, negative control of f-2810403D21Rik/Mirf. MirNC, negative control of Mir26a. (B) A confocal assay showed the anti-autophagic effects of f-2810403D21Rik/Mirf in NMCMs and could be abrogated by introduction of Mir26a. The data were obtained from 3 independent experiments and 10 cells were scored in each experiment. *p < 0.05, **p < 0.01. Scramble, negative control of f-2810403D21Rik/Mirf. MirNC, negative control of Mir26a. (C) Forced expression of f-2810403D21Rik/Mirf disrupts the expression of autophagy-related proteins by regulation of Mir26a. n = 6. *p < 0.05 vs. Scramble; #p < 0.05 vs. f-2810403D21Rik/Mirf. Scramble, negative control of f-2810403D21Rik/Mirf. MirNC, negative control of Mir26a.
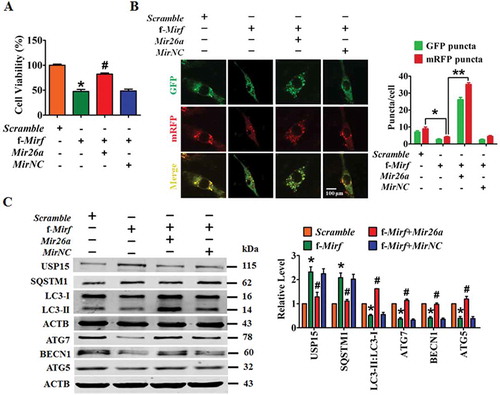
Figure 8. Silencing of 2810403D21Rik/Mirf alleviates myocardial injury and protects heart function in a mouse model of MI. (A) Upregulation of Mir26a in the hearts of WT mice after inhibition of 2810403D21Rik/Mirf. n = 6. **p < 0.01 vs. AAV9-sh-Scr. AAV9-sh-Scr, negative control of AAV9-sh-2810403D21Rik/Mirf. Silencing of 2810403D21Rik/Mirf by AAV9-sh-2810403D21Rik/Mirf improved heart function (B) (n = 11. *p < 0.05), reduced infarct size (C) (n = 5. **p < 0.01 vs. Sham; ##p < 0.01 vs. MI) in MI mice. Transmission electron microscopy (D) (n = 5. **p < 0.01) and western blot assay (E) (n = 6. *p < 0.05 vs. Sham; #p < 0.05 vs. MI) show the pro-autophagic effects of 2810403D21Rik/Mirf inhibition. The yellow arrows point to autophagic vesicles. AAV9-sh-Scr, negative control of AAV9-sh-2810403D21Rik/Mirf. (F) Proposed autophagic signaling mechanism for 2810403D21Rik/Mirf and Mir26a during MI. Cardiac stress inhibits expression of Mir26a by increasing the expression of 2810403D21Rik/Mirf and blocks autophagy, resulting in post-transcriptional de-repression of USP15 and leading to ischemic injury and cardiac dysfunction, respectively.
Table 1. The primers in RT-qPCR assays.
