Figures & data
Table 1. Brief description of receptors for organelle-specific autophagy
Table 2. The protective effects of organelle-specific autophagy on various inflammatory diseases
Figure 1. Quality control of multiple organelles by organelle-specific autophagy. (A) Mitophagy is of great importance in maintaining functional homeostasis of mitochondria, which is initiated by PINK1-PRKN-dependent and independent pathways. It requires TOMM and TIMM23 for the import and subsequent cleavage of PINK1 in functional mitochondria, while its damage and dysfunction are followed with PINK1 accumulation on the OMM. VDAC, RHOT1, MFN1/2 proteins act as phosphoubiquitin substrates for PINK1- and PRKN-dependent mitophagy. Outer mitochondrial membrane (OMM) proteins, including BNIP3L, FUNDC1, BNIP3, AMBRA1, BCL2LI3, FKBP8, CHDH, and DISC1, can detect damaged mitochondria and mediate mitophagy by interacting with MAP1LC3B protein directly. In the pathogenesis of mitochondrial dysfunction, IMM proteins, such as PHB2 and cardiolipin, are responsible for the initiation of mitophagy after translocation from IMM to OMM. Additionally, several post-transcriptional modification mechanisms are involved in regulating mitophagy: SIAH1, MUL1 and ARIH1 function as E3 ubiquitin ligases that target OMM proteins, and TBK1 can enhance mitophagy by phosphorylating autophagic receptors. (B) Nucleophagy is programmed for selective removal of nuclear components through the process of autophagy. LMNB1 and chromatin can be degraded via autophagic machinery under senescence exposure. (C) Four receptors reportedly contribute to sequestration of isolated cargos of ER into autophagosomes, including RETREG1, SEC62, CCPG1, and RTN3, which are essential for initiation of reticulophagy after binding with MAP1LC3B. Both RETREG1 and SEC62 are primarily responsible for the turnover of ER sheets, while CCGP1 and RTN3 for the ER-tubules. (D) Pexophagy is deemed great potential in quality control of peroxisome and might be induced by following mechanisms: overexpression of PEX3, activation of ATG9A-TNKS/TNKS2-PEX14 complex, phosphorylation and mono-ubiquitination of PEX5 via ATM and the PEX2-PEX10-PEX12 complex, which recognized by SQSTM1, as well as ABCD3-dependent NBR1-MAP1LC3B pathway in overexpression of PEX2. (E) NUFIP1 serves as the major receptor for ribophagy machinery via specific binding to MAP1LC3B in the assist of ZNHIT3. USP10 and G3BP1 are found to be the mammalian homologs of ribophagy receptors in yeast, suggesting their potential roles in mammal ribophagy. (F) Lysophagy is indispensable for quality control of lysosomes. LGALS3 can sense damaged lysosomes and further recruit TRIM16 to initiate autophagic machinery by ubiquitinating autophagy associated molecules, including ULK1 and ATG16L1. In addition, FBXO27 can serve as a ubiquitinating glycoprotein, which regulate the recruitment of autophagic machinery in SQSTM1-MAP1LC3B pathway
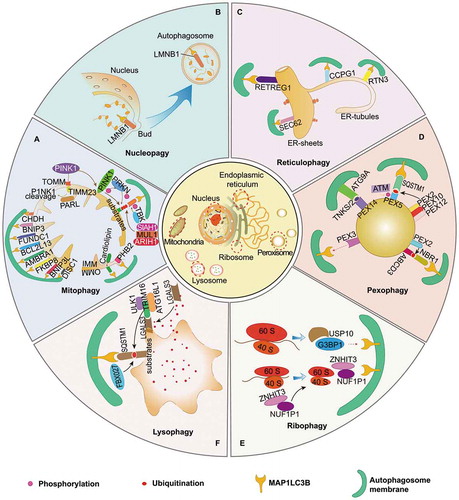
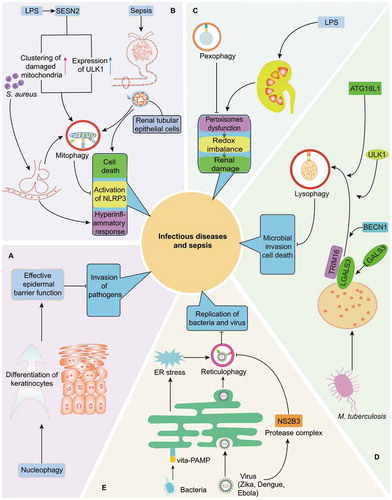
Figure 2. Quality control of multiple organelles through organelle-specific autophagy in infection and sepsis. (A) Nucleophagy is critically involved in preventing the invasion of pathogens by maintaining the integrity of epidermal barrier. (B) As initiated by LPS induced SESN2 upregulation, S. aureus-induced pneumonia as well as sepsis-related renal dysfunction, mitophagy is essential for the balance of inflammatory response and survival of cells in infection or septic challenge by limiting persistent NLRP3 inflammasome activation and eliminating damaged mitochondria respectively. (C) Induction of pexophagy attenuates LPS-mediated renal damage by restoring dysfunction of peroxisomes and redox imbalance. (D) Induction of lysophagy reportedly limits the invasion of intracellular M. tuberculosis in LGALS3- and TRIM16-dependent pathways, accompanied with recruitment of core autophagy proteins, including BECN1, ULK1 and ATG16L1. (E) Reticulophagy can promote the elimination of bacteria and virus via following mechanisms: resolving ER stress under exposure of Gram-positive bacteria and directly limiting replication of Zika, Dengue, and Ebola viruses. However, reticulophagy can be suppressed by NS2B3, a virus protease complex that may cleave RETREG1 within RHD domain