Figures & data
Figure 1. SUMOylation of PDPK1. (A) PDPK1 with a molecular weight of 210 kDa in the brain of mice. Lysates from the spleen, lung, kidney and brain of BALB/c mice were performed by in vivo SUMOylation detection assay described in material and method section and WB assay with using indicated antibodies. (B) PDPK1*S levels in the brain of drug-treated mice. Mice were intraperitoneally injected with rapamycin (0.4 mg/kg), LY294002 (1.5 mg/kg), INS (1 IU/kg), or DMSO. Lysates from the spleen, lung, kidney and brain of BALB/c mice were performed by in vivo SUMOylation detection assay described in material and method section and WB assay with using indicated antibodies. (C and D) PDPK1*S levels in wild-type and PDPK1-transfected cells. HEK293T cells and FLAG-PDPK1 transfected HEK293T cells were separately treated with rapamycin (R, 5 μM), LY294002 (L, 10 μg/ml), INS (I, 100 nM), or DMSO (D) for 24 h followed by in vivo SUMOylation detection assay described in material and method section and WB assay with using indicated antibodies (# indicates nonspecific bands). (E) The 210-kDa form of PDPK1 was formed upon RNA or DNA virus infections. RNA virus infections included IBDV infection (MOI = 10) in DF-1 cells for 7 h, influenza virus H7N9 (MOI = 10) and PR8 (MOI = 8) infections in A549 cells for 9 h, and VSV infection (MOI = 10) in HEK293T cells for 9 h. DNA virus infections contained PCV2 (MOI = 10) and PRV (MOI = 1) infections in PK15 cells for 72 h. The resultant samples are followed an in vivo SUMOylation detection described in material and method section with using indicated antibodies in immunoblotting analysis (# indicates nonspecific bands). (F) HEK293T cells co-transfected with FLAG-PDPK1 and PCI-NEO-UBE2I and empty vector HA, HA-SUMO1, HA-SUMO2, or HA-SUMO3 for 24 h is followed an in vivo SUMOylation detection by using indicated antibodies in immunoblotting analysis. (G) SUMOylation of PDPK1 is detected with indicated antibodies in immunoblotting analysis, after HEK293T cells were co-transfected with FLAG-PDPK1 and ShCon. or ShUBE2I for 20 h and then treated with INS for 24 h. (H) Colocalization between PDPK1 and SUMO1 in A549 cell lines. Scale bar: 10 μm. (I) HEK293T cells were co-transfected with PCI-NEO-UBE2I, HA-SUMO1 and empty vector FLAG, FLAG-SENP1, FLAG-SENP3, or MYC-PDPK1 for 24 h, and in vivo SUMOylation (described in material and method section) of PDPK1 is detected with using indicated antibodies in immunoblotting assay. (J) Map of SUMOylation sites and SUMO interaction sites in alignment of PDPK1 protein sequence from different species. (K) HEK293T cells were co-transfected with PCI-NEO-UBE2I and FLAG-PDPK1, FLAG-PDPK1CD, FLAG-PDPK1ID, FLAG-PDPK1SD, MYC-SUMO1, or empty vector MYC for 48 h. Co-IP assays were performed by using indicated antibodies. (L) HEK293T cells were co-transfected with PCI-NEO-UBE2I, MYC-SUMO1 and FLAG-PDPK1 or FLAG-PDPK1CD, FLAG-PDPK1ID, empty vector FLAG for 48 h, respectively. Co-IP assays with indicated antibodies. (M) HIS-PDPK1 and HIS-PIK3C3 were expressed in BL21 and purified. In vitro SUMOylation assay was then performed. Immunoblotting analysis of PDPK1 by using indicated antibodies. (N) SUMOylation analysis of PDPK1 by nickel beads affinity-isolation assay under denaturing condition. *, p < 0.05; **, p < 0.01; ***, p < 0.001
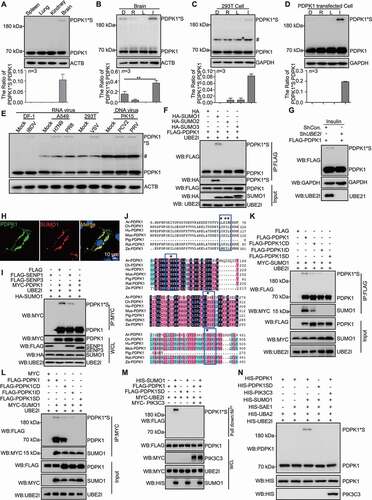
Figure 2. SUMOylation of PDPK1 is critical for activating the PDPK1-AKT1-MTOR pathway. (A) Phosphorylation analysis of RPS6KB1, AKT1, MTOR and PDPK1 in PDPK1 KD cells. PDPK1 KD HEK293T cells were separately transfected with vectors expressing FLAG-PDPK1, FLAG-PDPK1CD, FLAG-PDPK1ID, or FLAG-PDPK1SD for 48 h. Cell lysates were immunoblotted using indicating antibodies. (B) Vectors expressing FLAG-PDPK1, FLAG-PDPK1-SUMO1 (FLAG-P-S1), and FLAG-PDPK1SD-SUMO1 (FLAG-PSD-S1) were separately transfected into PDPK1 knocked down cells. The resultant cells were then lysed and subjected to immunoblotting analyses by using indicated antibodies. (C) PDPK1 KD HEK293T cells were transiently transfected with FLAG, FLAG-PDPK1, FLAG-PDPK1S241A, FLAG-PDPK1S241D, FLAG-PDPK1SD, FLAG-PDPK1SD and S241A, or FLAG-PDPK1SD and S241D for 48 h. Phosphorylation of MTOR, AKT1, and ULK1 is detected using indicated antibodies. *, p < 0.05; **, p < 0.01; ***, p < 0.001
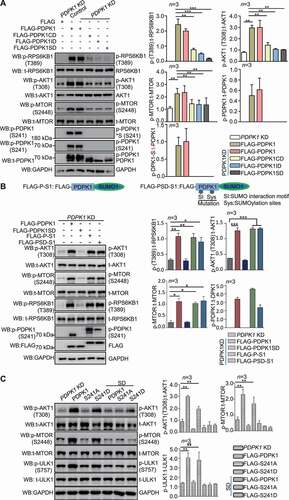
Figure 3. SUMOylation of PDPK1 is critical for regulating autophagy flux. Expression of PDPK1, PDPK1S241A, PDPK1S241D, PDPK1SD, PDPK1SD and S241A and PDPK1SD and S241D in PDPK1 knockdown cells treated with (A) or without (B) of CQ (50 μM) for 4 h. Cell lysis samples were subjected to immunoblotting analysis by using indicated antibodies. *, p < 0.05; **, p < 0.01; ***, p < 0.001
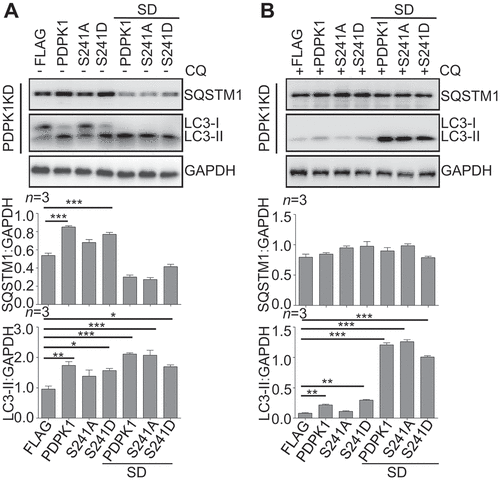
Figure 4. SUMOylation of PDPK1 is required for its interaction with PIK3C3. (A) The colocalization analysis between PDPK1 and PIK3C3. A549 cells were transiently transfected with FLAG-PDPK1 and MYC-PIK3C3 for 48 h, the cells were fixed and immunostained with anti-FLAG and anti-MYC antibodies and then observed under super-resolution microscope (SIM). Scale Bar: 2 μm. (B) Endogenous PDPK1 interaction with PIK3C3. 293 T cells were lysed and then performed anti-PIK3C3 IP analysis of endogenous PDPK1 and PIK3C3 by using indicated antibodies. (C) Verification of direct interaction between PIK3C3 and PDPK1 in GST-affinity-isolation assay by using recombinant HIS-PDPK1 and GST-PIK3C3 or GST proteins purified from E.coli BL21. (D) SUMOylated and phosphorylated PDPK1 interacted strongly with PIK3C3. HEK293T cells were transiently transfected with FLAG, FLAG-PDPK1, FLAG-PDPK1S241A, FLAG-PDPK1S241D, FLAG-PDPK1SD, FLAG-PDPK1SD and S241A, or FLAG-PDPK1SD and S241D together with MYC-PIK3C3 for 48 h. Anti-FLAG IP analysis was conducted to detect MYC-PIK3C3, FLAG-PDPK1, and phosphorylated PDPK1. (E) Binding of PDPK1 to PIK3C3 was enhanced in rapamycin- but not in INS-treated cells. HEK293T cells were transiently co-transfected with FLAG-PDPK1 and PIK3C3 for 24 h. The resultant cells were treated with rapamycin (R, 5 μM), INS (I, 100 nM), or DMSO (D) for another 24 h, respectively. Anti-FLAG IP of FLAG-PDPK1 and PIK3C3 is performed by using indicated antibodies. (F) Mapping interaction domain of PIK3C3 with PDPK1. Co-IP and immunoblotting assays were performed with anti-MYC and anti-FLAG antibodies in extracts of HEK293T cells transfected with MYC-PDPK1 together with empty vector FLAG, FLAG-PIK3C3, FLAG-SH2, FLAG-Helical, or FLAG-Kinase for 48 h
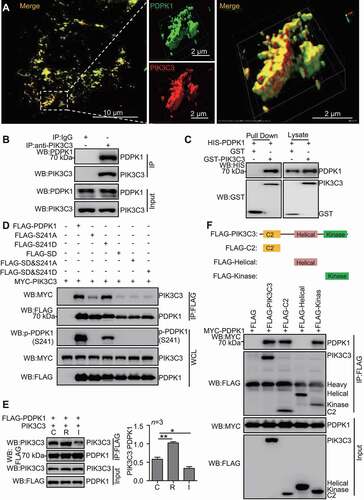
Figure 5. PIK3C3 inhibits SUMOylation of PDPK1. (A) PIK3C3 overexpression downregulated SUMOylated PDPK1. HEK293T cells were transiently co-transfected with MYC-PIK3C3 together with FLAG-PDPK1, PCI-UBE2I, and HA-SUMO1 for 48 h. FLAG precipitation and immunoblot analyses of FLAG-PDPK1 were performed by using indicated antibodies. (B) PIK3C3 deletion promoted SUMOylation of PDPK1. PIK3C3 KO or WT HEK293T cells were co-transfected with HA-SUMO1 and PCI-UBE2I together with FLAG-PDPK1 for 48 h. Anti-FLAG IP and immunoblot analyses were performed by using indicated antibodies. (C) PDPK1 SUMOylation is inhibited by PIK3C3. HEK293T cells were transiently co-transfected with FLAG-PDPK1 and PCI-UBE2I together with increasing amounts of MYC-PIK3C3 for 48 h. Anti-IgG or -FLAG IP and immunoblot analyses were performed by using indicated antibodies. (D) HEK293T cells were transiently co-transfected with FLAG-PDPK1 and MYC-SENP3 together with or without HA-PIK3C3 for 48 h. Anti-IgG or -FLAG IP and immunoblotting analyses were conducted by using indicated antibodies. (E) The expression of PIK3C3 inhibited phosphorylation of PDPK1, AKT1 and MTOR. HEK293T cells were transiently transfected with empty vector or FLAG-PIK3C3 for 48 h. Cell lysates were subjected to western blotting with indicated antibodies. (F) Knockout of PIK3C3 increased the phosphorylation of PDPK1, AKT1 and MTOR. Lysates of WT and PIK3C3-null HEK293T cells was subjected to immunoblot analysis by using indicated antibodies
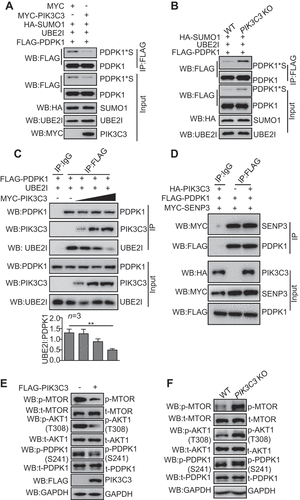
Figure 6. NonSUMOylated PDPK1 located to the ER facilitates autophagosome biogenesis. (A) The ER co-localization with nonSUMOylated PDPK1. 293 T cells were transiently co-transfected with DsRed-ER and FLAG-PDPK1, FLAG-PDPK1S241D, FLAG-PDPK1S241A, FLAG-PDPK1SD, FLAG-PDPK1SD and S241A, or FLAG-PDPK1SD and S241D for 48 h. Cells were immunostained with anti-FLAG antibodies and observed under a confocal microscope. Scale bar: 10 μm. (B) Live-cell time-lapse images of Vero cells expressing PDPK1 (blue), GFP-LC3 (green), and DsRed-ER (red). Cells were observed under a Nikon A1R/A1 laser-scanning confocal microscope. White arrowheads indicate LC3 co-localization with PDPK1. Scale bar: 2 μm. (C) Three-dimensional image of PDPK1 co-localization with LC3 and the ER. HEK293T cells were transiently co-transfected with DsRed2-ER, GFP-LC3, or FLAG-PDPK1 for 24 h, and twenty-one cells immunostained with anti-FLAG antibody were observed by SIM from each treatment. Scale Bar: 0.5 μm. (D) Quantitative analysis of ER-PDPK1-LC3 structures or ER-LC3 structures relative to LC3 puncta in (C). Results is shown as mean ± SD. (E) LC3-II and phosphorylated PDPK1 were main modified isoforms in the subcellular ER fraction. ER was obtained from HEK293T cells by using the ER0100 ER isolation kit and then subjected to immunoblot analysis by using indicated antibodies. (F) NonSUMOylated PDPK1 interacted with endogenous LC3-II, WIPI2, as well as ATG12-ATG5. HEK293T cells were transiently transfected with FLAG, FLAG-PDPK1, FLAG-PDPK1S241D, FLAG-PDPK1S241A, FLAG-PDPK1SD, FLAG-PDPK1SD and S241A, or FLAG-PDPK1SD and S241D for 48 h. Anti-FLAG IP and immunoblotting analyses were performed by using indicated antibodies
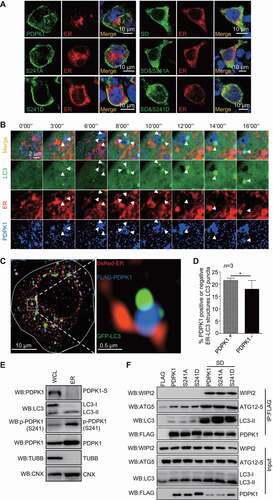
Figure 7. NonSUMOylation of PDPK1 is critical for lipidation of LC3. (A) Live-cell time-lapse images of Vero cells expressing PDPK1 (blue), GFP-LC3 (green), and DsRed-SQSTM1 (red) for 24 h. Cells were observed under a Nikon A1R/A1 laser-scanning confocal microscope. (B) Three-dimensional dynamic image of autophagosome formation. HEK293T cells were transiently transfected with FLAG-PDPK1, GFP-LC3, and DsRed-SQSTM1 for 24 h and then observed by SIM. Fifty cells in each treatment were recorded. (C) Immunoblot analysis of lysates from WT-PDPK1, PDPK1 KD, or PIK3C3 KO HEK293T cell lines in the presence or absence of CQ by using indicated antibodies. (D) Immunoblot analysis of lysates from WT-PDPK1 or PDPK1 KD HEK293T cell lines overexpressing FLAG or FLAG-PDPK1 in presence or absence of CQ by using indicated antibodies
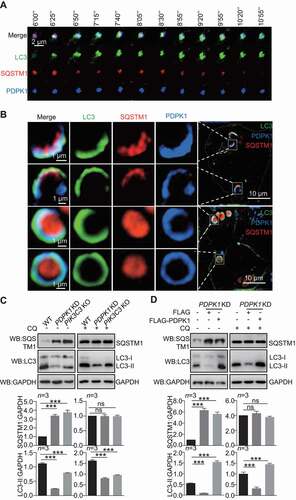
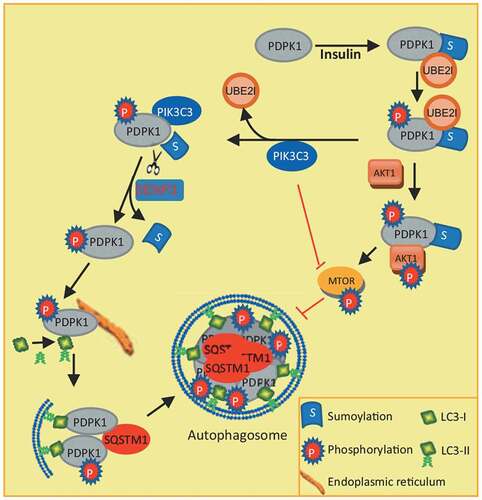
Figure 8. Model for how PDPK1 regulates autophagosome formation. SUMOylation of PDPK1 induces its phosphorylation. Subsequently, SUMOylated and phosphorylated PDPK1 activates the AKT1-MTOR pathway and negatively regulates autophagy. In turn, the PDPK1-AKT1-MTOR pathway is inactivated by PIK3C3 binding to SUMOylated and phosphorylated PDPK1. PIK3C3 results in deSUMOylation of PDPK1 by disrupting PDPK1 interaction with UBE2I. NonSUMOylated PDPK1 links LC3 to the ER subdomain and promotes the lipidation of LC3. Subsequently, nonSUMOylated PDPK1 facilitates the expansion of the phagophore to form the enclosed autophagic vacuole