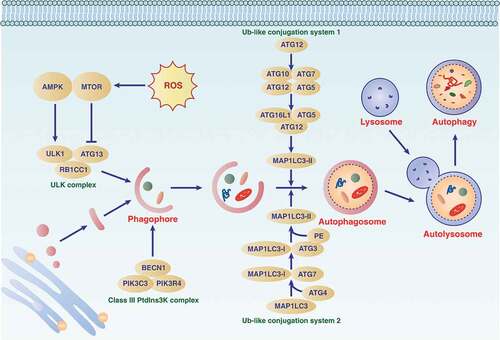
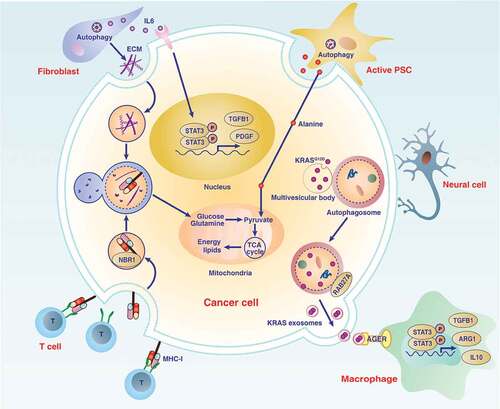
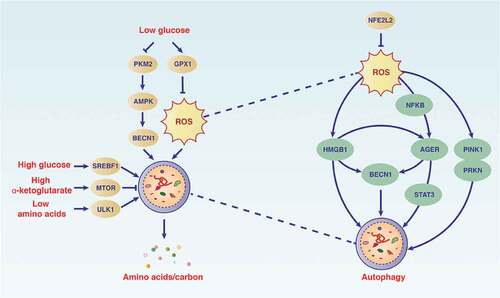
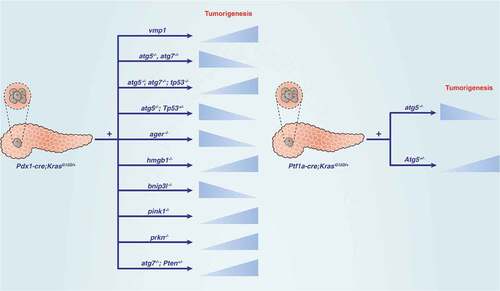
Figures & data
Table 1. Monotherapies that affect autophagy in PDAC
Table 2. Combination therapies that affect autophagy in PDAC
Table 3. Published clinical studies on using HCQ to inhibit autophagy in PDAC