Figures & data
Figure 1. Lack of CREG1 remarkedly reduced in endurance performance in 9-month-old mice. (A and E) Exercise performance was assessed in age-matched Creg1fl/fl and creg1;Ckm-Cre mice by performing a treadmill exhaustion test. Time to exhaustion (A) and corresponding quantitative analysis (B); running distance (C) and corresponding quantitative analysis (D); and work (E) performed during the test. (Creg1fl/fl 3-month-old: n = 6, creg1;Ckm-Cre 3-month-old age: n = 7, Creg1fl/fl 9-month-old: n = 6, creg1;Ckm-Cre 9-month-old age: n = 6). (F and G) Exercise performance was also assessed in 9-month-old creg1;Ckm-Cre and reCREG1-treated mice by performing a treadmill exhaustion test. Quantitative analysis of time to exhaustion (F). Quantitative analysis of running distance (n = 6) (G). Data are shown as mean ± SEM, **p < 0.01. Creg1fl/fl: mice bearing Creg1-floxed alleles; creg1;Ckm-Cre: skeletal muscle-specific knockout creg1 mice; reCREG1: treated with recombinant CREG1 protein
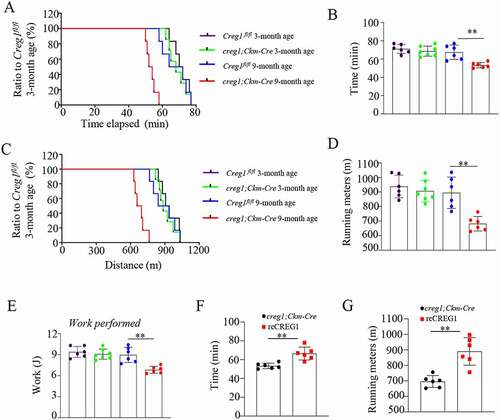
Figure 2. Creg1 ablation reduced the distribution of type I fiber and modulated the skeletal muscle mitochondrial morphology in 9-month-old creg1;Ckm-Cre mice. (A and B) Representative photomicrographs used for quantitative analysis of immunostaining for MYH1/MHC-I in the gastrocnemius and soleus of 9-month-old creg1;Ckm-Cre mice. Type II fibers were negative to antibody anti-MYH1. The analysis was performed using 5 fields per section (50 μm). An average of 400 fibers was analyzed in the gastrocnemius and soleus for each mouse. (C) RT-PCR analysis of Myh isoforms (Myh1, Myh2/Mhc2a and Myh4/Mhc2b) expression in the gastrocnemius. (D) Electron microscopy (EM) analysis of gastrocnemius from 9-month-old Creg1fl/fl and 9-month-old creg1;Ckm-Cre, scale bars: 1.0 μm. Size (E), mitochondrial number (F) and the ratio of abnormal mitochondria (G) in the two groups. (H) MtDNA analysis of gastrocnemius in the two groups (n = 3). (I and J) Western blotting analysis of mitochondrial marker proteins in the gastrocnemius of 9-month-old Creg1fl/fl and 9-month-old creg1;Ckm-Cre, PTGS2, COX4I1, and TOMM20 expression. (K) Confocal microscopy analysis of the localization of his-CREG1 and MYH1, scale bars: 50 μm. (L and M) Western blotting analysis of mitochondrial marker proteins in the gastrocnemius of 9-month-old creg1;Ckm-Cre treated with saline and treated with recombinant (re)CREG1 protein (reCREG1), PTGS2, COX4I1, and TOMM20 expression. (N) Electron microscopy (EM) analysis of gastrocnemius from 9-month-old creg1;Ckm-Cre treated or not treated with reCREG1, scale bars: 1.0 μm. (O) Size, mitochondrial number (P) and the ratio of abnormal mitochondria (Q) in the two groups. Yellow asterisks represented normal mitochondria. Red asterisks represented abnormal mitochondria. Data are shown as mean ± SEM, *p < 0.05, **p < 0.01
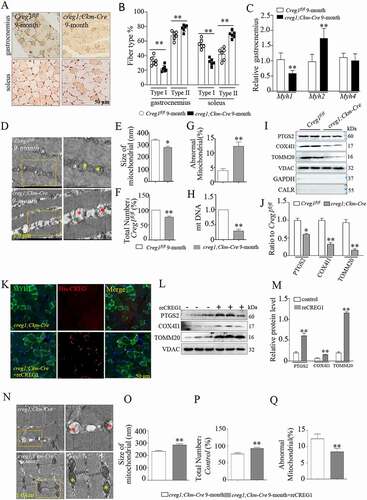
Figure 3. Deficiency of CREG1 enhanced mitophagy in the gastrocnemius of 9-month-old creg1;Ckm-Cre mice. (A and C) Western blotting (A) and quantification analysis showed the expression of PINK1 (B) and PRKN (C) in the mitochondria of the gastrocnemius of 3- and 9-month-old Creg1fl/fl and creg1;Ckm-Cre mice. (D and F) Western blotting (D) and quantification analysis (E and F) showed the expression of LC3-II and SQSTM1 protein in the mitochondria of the gastrocnemius of 3- and 9-month-old Creg1fl/fl and creg1;Ckm-Cre mice. (G) Confocal microscopy analysis of the colocalization of LC3B and TOMM20, yellow asterisks represented the colocalization of LC3B and TOMM20, scale bars: 50 μm. (H and I) Western blotting (H) and quantification analysis (I) showed the expression of SQSTM1 and LC3-II protein in the mitochondria of the gastrocnemius treated with colchicine (0.4 mg/kg·day). Data are shown as mean ± SEM, **p < 0.01, ##p < 0.01
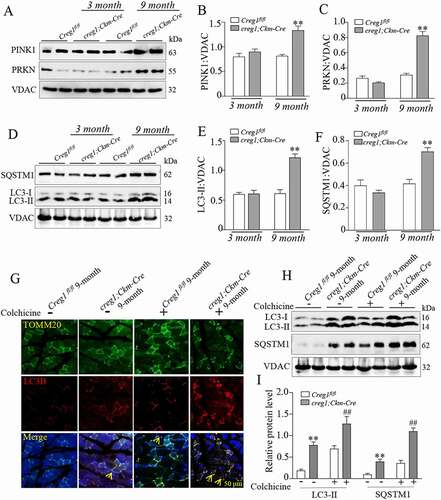
Figure 4. CREG1 altered the morphology and function of mitochondria in C2C12 cells. (A) MitoTracker Red staining revealed the morphology of mitochondria in the AdGFP, AdCREG1, siScramble, and siCreg1-treated C2C12 cells. Scale bars, left column: 20 μm. Right column, magnified view of the area in the image from the left column indicated with a yellow square. Yellow arrows pointed out mitochondria. (B and D) Electron microscopy (EM) analyzed the morphology of mitochondria in the AdGFP, AdCREG1, siScramble, and siCreg1-treated C2C12 cells, left column, scale bars: 1.0 μm. Right column, magnified view of the area marked with a yellow square. Quantification for mitochondrial number (C) and the ratio of abnormal mitochondria (D). (E) MtDNA analysis in the AdGFP, AdCREG1, siScramble, and siCreg1-treated C2C12 cells. Yellow asterisks represented normal mitochondria. Red asterisks represented abnormal mitochondria. (F and G) Western blotting (F) and quantification analysis (G) revealed the expression of PTGS2, COX4I1, and TOMM20 in mitochondrial fraction of AdGFP, AdCREG1, siScramble, and siCreg1-treated C2C12 cells. (H and I) Western blotting (H) and quantification analysis (I) revealed the expression of DNM1L, MFF, OPA1 and MFN2 in mitochondrial fraction of AdGFP, AdCREG1, siScramble and siCreg1-treated C2C12 cells. Representative oxygen consumption curves (J) and quantification analysis (K) in the AdGFP, AdCREG1, siScramble, and siCreg1-treated C2C12 cells. Basal respiration rate was measured followed by proton leak after the addition of oligomycin, maximal respiration after the addition of FCCP, and non-mitochondrial respiration after the addition of rotenone. Data are shown as mean ± SEM, n = 3, *p < 0.05, **p < 0.01, ##p < 0.01
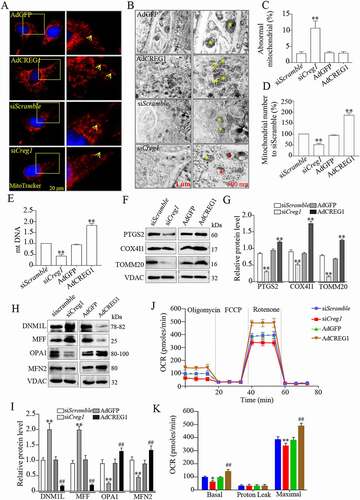
Figure 5. Deficiency of CREG impaired the mitophagy in C2C12 cells. (A and B) Western blotting analysis to measure the expression of PINK1, PRKN, LC3-II and SQSTM1 in the mitochondria from the siScramble and siCreg1 cell groups. (C) Confocal images of colocalization of GFP-LC3B puncta and mitochondria (red-MitoTracker) in C2C12 cells transfected with the Creg1 siRNA during growth in CCCP (10 μM) for 1 h. Scale bar: 10 μm. (D) Quantification of colocalization of GFP-LC3B puncta and mitochondria. n = 3. (E and F) Western blotting (E) and quantification analysis (F) showed the expression of LC3-II and SQSTM1 protein in the mitochondria of the gastrocnemius treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP, 10 μΜ, 1 h). Data are shown as mean ± SEM, **p < 0.01, ##p < 0.01
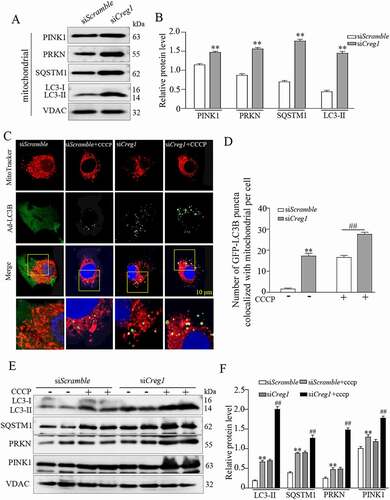
Figure 6. Interaction between CREG1 and HSPD1 in vitro and in vivo. (A and B) 293 T cells were transfected with Creg1-HA plasmid and Hspd1-FLAG plasmid together. Co-IP of CREG1 and HSPD1 in 293 T cells. (C) Confocal microscopy analysis of the colocalization of CREG1-GFP and HSPD1, scale bars: 20 μm. (D and E) Co-IP assay showed the interaction between CREG1 and HSPD1 in the skeletal muscle protein from 9-month-old creg1;Ckm-Cre and Creg1fl/fl mice. (F and G) Western blotting and quantification analysis showed the expression of HSPD1 protein in the skeletal muscles of 3-month or 9-month-old mice Creg1fl/fl and creg1;Ckm-Cre mice. (H and I) Western blotting analysis demonstrated the expression of HSPD1 protein in the skeletal muscles of 9-month-old creg1;Ckm-Cre with or without reCREG1 protein treatment. (J and K) Western blotting and quantification analysis showed the expression of HSPD1 protein in siScramble, siCreg1, and siCreg1+ MG132 (10 μmol/L) cells. (L and M) Western blotting and quantification analysis showed the expression of CREG1 protein in siScramble, siHspd1, and siHspd1+ MG132 (10 μmol/L) cells. Data are shown as mean ± SEM, **p < 0.01, &p < 0.01
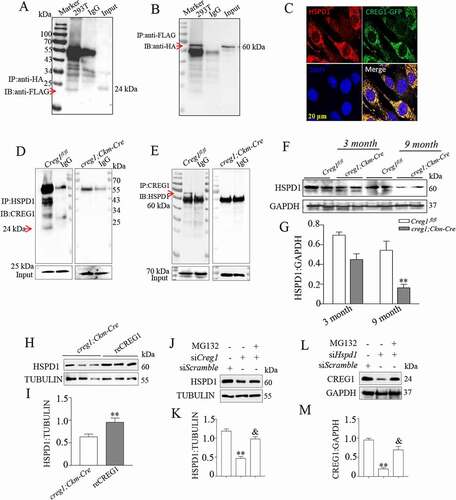
Figure 7. CREG1 directly interacted with the peptide fragment 401–573 aa of HSPD1. (A and C) Co-IP analysis to detect interaction between CREG1 and peptide fragments 1–200 aa (A), 201–400 aa (B), and 401–573 aa (C) of HSPD1. IP antibody: anti-FLAG, IB antibody: anti-CREG1. Input: CREG1. (D and E) Co-IP analysis to detect interaction between HSPD1 with different peptide fragments of CREG1: 1–100 aa (D) and 130–220 aa (E). IP antibody: anti-HIS, IB antibody: anti-HSPD1. Input: HSPD1
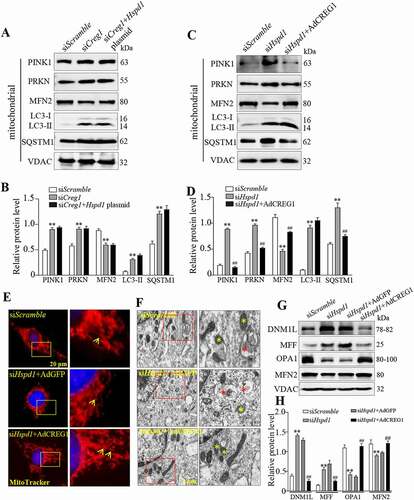
Figure 8. Overexpression of CREG1 rescued siHspd1-induced mitophagy in C2C12. (A and B) Western blotting analysis showed the expression of PINK1, PRKN, MFN2, LC3-I/LC3-II and SQSTM1 protein in siScramble, siCreg1, and siCreg1+ Hspd1 plasmid cells. (C and D) Western blotting analysis showed the expression of PINK1, PRKN, MFN2, LC3-I/LC3-II and SQSTM1 protein in the siScramble, siHspd1, and siHspd1+ AdCREG1 cells. (E) MitoTracker Red staining revealed the expression of mitochondria in siScramble, siHspd1+ AdGFP and siHspd1+ AdCREG1 three C2C12 cells, scale bars: 20 μm. (F) Electron microscopy (EM) analysis of cells from the three groups, scale bars: 1.0 μm. Yellow asterisks represented normal mitochondria, red asterisks represented abnormal mitochondria. (G and H) Western blotting analysis to measure the expression of DNM1L, MFF, OPA1 and MFN2 in the mitochondria from the cell groups. Data are shown as mean ± SEM, **p < 0.01, ##p < 0.01