Figures & data
Figure 1. FYCO1 interacts with and is cleaved by activated CASP8. (A) Scatterplot representing proteins enriched upon CASP8 affinity isolation by mass spectrometry analysis. HeLa treated with TNFSF10/TRAIL were compared with untreated cells. The cut off for significant enrichment was set at 4-fold label-free quantitation (LFQ) values and a p-value of < 0.01 (-log (p-value) > 2). (B) WB of lysates and anti-CASP8 co-IP of HeLa cells treated with TNFSF10/TRAIL (500 ng/ml) and CHX (1 µg/ml) for the indicated timeframes. The WB was performed for the proteins indicated. (C) In vitro CASP8 cleavage assay. Coomassie Brilliant Blue staining of recombinant FYCO1-V5 followed by incubation with 1 U recombinant CASP8. Below, the WB for anti-V5. (D) In vitro CASP8 and CASP3 cleavage assays. Coomassie blue staining of recombinant FYCO1-V5 followed by incubation for 1 h with a titration of recombinant caspase units. Below, the WB for the antibodies indicated. (E) WB of HeLa lysates treated with TNFSF10/TRAIL (1 µg/ml, 2 h) in combination with different caspase inhibitors (20 µM).
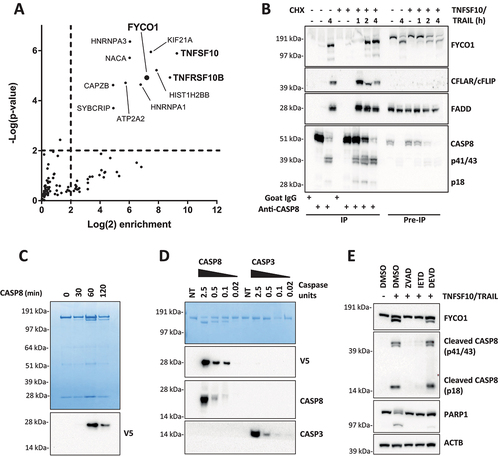
Figure 2. FYCO1 KD sensitizes cells to basal and DR-induced apoptosis. (A) HeLa cells were transduced with 4 different shRNAs for FYCO1 (#14; #15; #17 and #18) and control. Lysates were prepared after 3 or 7 days and WB analyses for the indicated proteins were performed. (B) FYCO1 expression in HeLa cells was knocked down with 2 different inducible shRNAs (ind. sh#14 and #17) by the addition of doxycycline (250 ng/ml) for the timeframes shown. Afterwards, WB analyses for the proteins indicated were performed. (C) WB to validate FYCO1 KD by 3 different siRNA oligos (40 nM, 72 h). (D) Cell viability assay (Cell titer Glo) of HeLa cells knocked down for FYCO1 by siRNA (40 nM, standard error of the average viability (n = 3). Statistical analysis was performed by two-way ANOVA with Dunnett test vs scramble control. (E) WB to validate FYCO1 knockout by CRISPR-Cas9 mediated gene editing. (F) Cell viability assay (Cell titer Glo) of control and FYCO1 knockout HeLa cells as in (E), subjected to titration of TNFSF10/TRAIL treatment for 24 h. Reported is the mean ± standard error of the average viability (n = 3). Statistical analysis was performed by two-way ANOVA with Dunnett test vs WT cells.
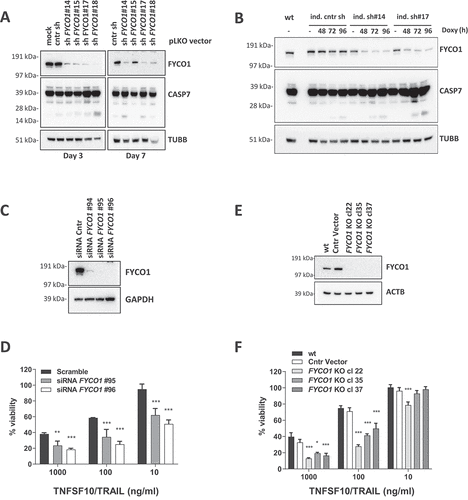
Figure 3. The absence of FYCO1 leads to the stabilization the DISC caused by impaired lysosomal degradation. (A) Cytosolic and membrane fractions of HeLa cells knocked out for FYCO1 by CRISPR-Cas9 and treated with TNFSF10/TRAIL (2 h, 500 ng/ml) were prepared. Anti-CASP8 co-IP was performed. Shown is the WB analysis for the proteins indicated. (B) Cytosolic and membrane fractions of HeLa cells transfected with scramble or siRNA for FYCO1 (#95, 40 nM, 72 h) treated or not with TNFSF10/TRAIL (500 ng/ml, 2 h) were prepared and anti-CASP8 co-IPs were performed. Shown are the WB tested for the proteins indicated. (C) Schematic representation of the DISC isolation. (D) WB analysis of the TNFSF10/TRAIL DISC. Cells were stimulated with 500 ng/ml FLAG-tagged TNFSF10/TRAIL for the indicated timeframes. The receptor-associated proteins were then immunoprecipitated with anti-FLAG agarose and WB analysis for the proteins indicated was performed. (E) TNFRSF10B/DR5 localization assay. Control and KO FYCO1 HeLa cells were treated with bafilomycin A1 (1 h, 100 nM) and then with TNFSF10/TRAIL (2 h, 1 µg/ml). Representative immunofluorescence showing TNFRSF10B/DR5 (red) and LAMP1 (green) staining (63X magnification). Colocalization is highlighted in yellow and represented by a scatter plot of pixels in the two fluorescence channels. The percentage of TNFRSF10B/DR5 colocalization with LAMP1 was calculated for 10 representative fields and used for box plot graphical representation and statistical analysis by Student t-test.
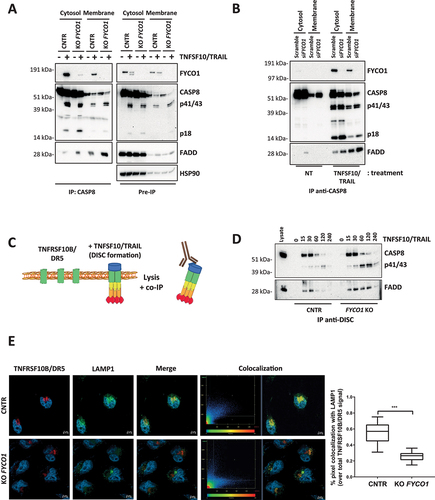
Figure 4. Cleavage of FYCO1 occurs at Aspartate 1306 and blunts its function. (A) Scheme of the FYCO1 protein structure. Arrows indicate two putative CASP8 cleavage sites. (B) In vitro CASP8 cleavage assay. Bead-bound FYCO1 WT-V5, FYCO1D1238A-V5, and FYCO1D1306A-V5 were incubated for 1 h with the indicated concentrations of recombinant CASP8. Then a WB detecting the V5 epitope was performed. (C) The TETD tetrapeptide is a preferential cleavage site for CASP8. The graph was obtained by plotting the normalized frequency scores of each aa at the CASP3 and CASP8 cleavage sites, as obtained by the SitePrediction web tool. (D) KO FYCO1 cells were transfected with empty pcDNA 3.1 vector or plasmids encoding FYCO1 WT, FYCO1D1238A, or FYCO1D1306A. Then cells were treated with TNFSF10/TRAIL (500 ng/ml, 2 h), lysed and a WB for the proteins indicated was performed. (E) HeLa KO FYCO1 were transfected with empty pcDNA 3.1 vector or plasmids encoding FYCO1 WT, or FYCO1D1306A. Then cells were treated with 500 ng/ml FLAG-tagged TNFSF10/TRAIL as indicated. Lysates were tested by WB for the indicated proteins. (F) Cells were treated as in (E) and the lysates were subjected to anti-DISC immunoprecipitation by anti-FLAG beads. Shown is a representative WB analysis for the protein indicated.
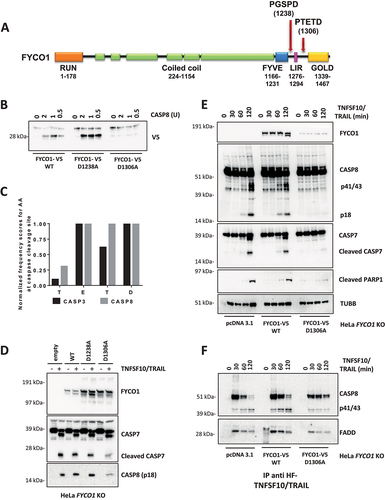
Figure 5. The C terminus of FYCO1 interacts with CCZ1. (A) Scheme of FYCO1 WT protein and of the truncated FYCO1 1–1306. (B) Cells were treated with 500 nM rapamycin (4 h), lysed and anti-FYCO1 or anti-rabbit IgG (isotype control) co-IPs were performed. Shown is a WB for the proteins indicated. (C) HeLa KO FYCO1 were reconstituted with either FYCO1 WT or its truncated version (1–1306), pre-treated with rapamycin as in (B), lysed and co-IP with anti-FYCO1 antibody was performed. Shown is the WB for the proteins indicated. (D) HeLa cells were transfected with the siRnas indicated (40 nM, 72 h), treated with 500 ng/ml TNFSF10/TRAIL, lysed, and WB for the proteins indicated was performed. (E) Lysates were prepared as in (D) and subjected to anti-DISC immunoprecipitation using anti-FLAG beads. The precipitated proteins were analyzed by WB with the antibodies indicated. (F) HeLa cells were transfected with two different siRNAs targeting FYCO1 and CCZ1. Cells were lysed after 48 and 72 h and WB analysis was performed for the indicated proteins. (G) TNFRSF10B/DR5 localization assay. HeLa cells were transfected with scramble, siRNA targeting FYCO1 (#96) or CCZ1 (#91) at 40 nM for 72 h. Afterward, they were pre-treated with bafilomycin A1 (1 h, 100 nM) and then with TNFSF10/TRAIL (2 h, 1 µg/ml). Representative immunofluorescence showing TNFRSF10B/DR5 (red) and LAMP1 (green) staining (63X magnification). Colocalization is highlighted in yellow and represented by a scatter plot of pixels in the two fluorescence channels. The percentage of TNFRSF10B/DR5 colocalization with lysosomes was calculated for 10 representative fields and used for box plot graphical representation and statistical analysis by Student t-test.
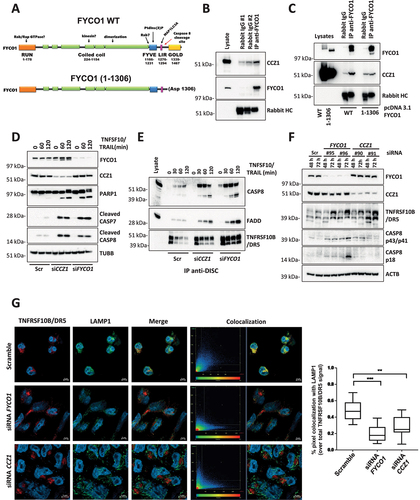
Figure 6. The C-terminal cleavage fragment of FYCO1 impairs FYCO1 binding to CCZ1. (A) Graphical scheme of FYCO1 C-terminal fragment (1307-end), V5-tagged and cloned into a doxycycline-inducible vector. (B) HeLa were transduced with either empty or FYCO1 1307-end encoding vector, induced by doxycycline (18 h, 250 ng/ml) and treated with the autophagy inducer rapamycin (500 nM, 4 h); then, anti-FYCO1 and rabbit IgG Co-IPs and WB for the indicated proteins was performed. (C) Samples were prepared as in (B). Anti-CCZ1 and control IgG1 Co-IPs and WB for the indicated proteins was performed. (D) HeLa transduced with either empty or FYCO1 1307-end encoding vector were induced for one or two days with doxycycline, in presence or not of Z-VAD-FMK (20 µM). Mean ± standard deviation of 4 experiments, statistical analysis by Two-way ANOVA with Tukey’s multiple comparison test. (E) Ectopic expression of FYCO1 C-terminal fragment was induced by doxycycline treatment (75 ng/ml, 12-24-36 h), then the cells were lysed and WB for the indicated proteins was performed. (F) Cells were treated as in (E). Then the samples were immunoprecipitated. The associated proteins were visualized by immunoblotting. (G) Receptor-associated proteins were isolated from cells depleted for FYCO1 expression by siRNA (#96) mediated KD for 48 and 72 h, then analyzed by immunoblotting.
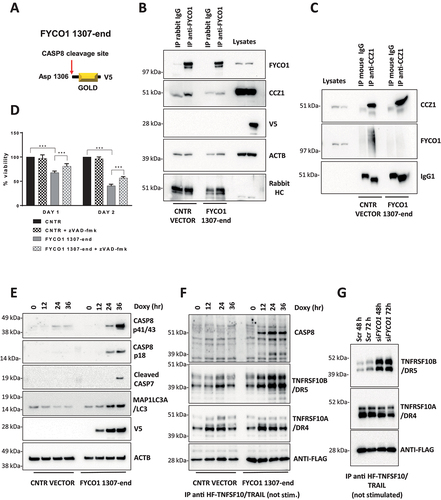
Figure 7. FYCO1 regulates TNF receptor complex turnover. (A) the TNFRSF1A/TNF-R complex was isolated from control and FYCO1 KO HeLa cells treated with TNF-Fc containing supernatant for different time points. Shown is the WB analysis for the proteins indicated. (B) WB analyses of the full cell lysates prepared as in (A). (C) WB analyses of the TNFRSF1A/TNF-R complexes obtained from FYCO1 KO HeLa cells transfected with empty vector, FYCO1 WT or its truncated (1–1306) mutant, treated with TNF-Fc containing supernatant for 5-15-30 min. (D) WB for p-NFKB1 levels in HeLa cells transfected with siRNAs against CCZ1 or FYCO1 (40 nM, 72 h). (E) Cells were treated as in (D) and then subjected to immunoprecipitation of the TNFRSF1A/TNF-R complex. Shown is an immunoblot for the proteins indicated. (F) Lysates of untreated and TNFSF10/TRAIL-treated cells (500 ng/ml, 4 h) were immunoprecipitated with ubiquitin-immobilized agarose and with control CL-4B Sepharose. Specific binding of FYCO1 and SQSTM1/p62 was visualized by WB. (G) WB analyses of anti-FYCO1 co-IPs in lysates of cells untreated or treated with TNF-Fc containing supernatant for 5 min.
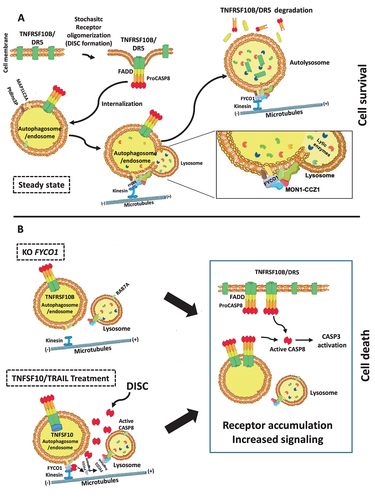
Figure 8. Proposed working model. (A) FYCO1 mediates efficient lysosome-mediated removal of stochastic CASP8 auto-activating complexes, and this is necessary to ensure cell survival. The fusion of endocytosed complexes with lysosomes is dependent on the interaction of the C-terminal GOLD domain of FYCO1 with the RAB7A GEF complex containing CCZ1-MON1A, further responsible for interaction with the HOPS complex. (B) In absence of FYCO1, the autolysosomal degradation of receptors is impaired and the accumulating receptors sensitize the cells for ligand-induced and constitutive apoptosis. Furthermore, if a certain stimulus threshold is reached (e.g., by addition of TNFSF10/TRAIL), CASP8-mediated cleavage of FYCO1 C-terminal domain prevents this lifeguard mechanism, thus allowing amplification of the apoptotic cascade and execution of cell death.