Figures & data
Table 1 Background information of MMC patients included in the HumanMethylation450 BeadChip and Sequenom EpiTYPER
Table 2 Methylation of the HOX genes using the HumanMethylation450 BeadChip and analysis
Figure 1. HOXB7 methylation studies by Sequenom EpiTYPER in MMC patients. (A) Localization of the studied amplicon (Chr17:46,685,144–46,685,550) within HOXB7 Exon 2. The amplicon covers 26 single CpGs and our assay provides data on 10 analytical CpG units. Nucleotide positions accord to the NCBI build 37/hg19. The CpG units studied by 450K Array (cg11041817, cg22622477 and cg07547765) and the in silico analysis (cg06493080, cg09357097) are also indicated. (B): Boxplot representing the methylation pattern of MMC patients and controls with box = 25th and 75th percentiles; bars = min and max values. The mean methylation level of each group is shown below the plot. (C): Methylation pattern for each CpG unit within the amplicon. Wilcoxon Rank-Sum test was performed. (D): Boxplot representing the methylation pattern of MMC patients and controls divided according to MTHFR 677C>T genotype with box = 25th and 75th percentiles; bars = min and max values. The mean methylation level of each group is shown below the plot.
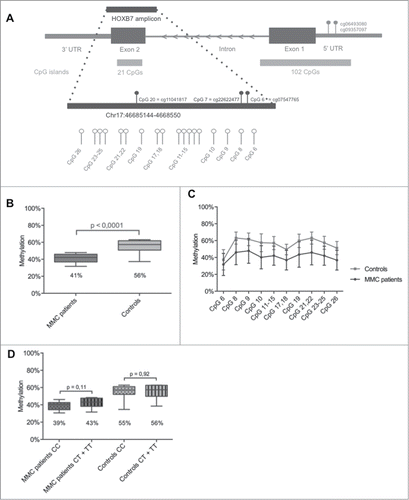
Figure 2. HOXB7 methylation studies by Sequenom EpiTYPER in pairs of unaffected siblings vs. MMC patients. (A) Boxplot representing the methylation pattern of affected siblings and unaffected siblings with box = 25th and 75th percentiles; bars = min and max values. The mean methylation level of each group is shown below the plot. (B) Methylation pattern for each CpG unit within the amplicon. Wilcoxon Rank-Sum test was performed.
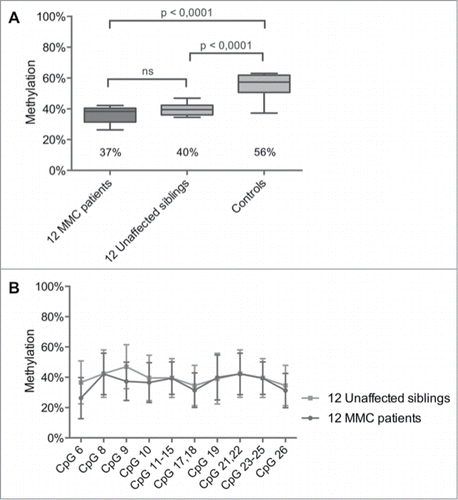
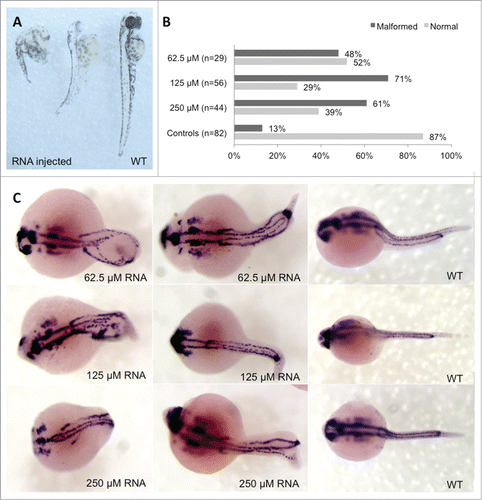
Figure 3. Phenotype analysis of Hoxb7a-overexpression in zebrafish embryos. (A) Phenotype analysis at 72 hpf of Hoxb7a mRNA injected zebrafish resulted in significant hypopigmentation and malformation in 66% of the injected zebrafish. These embryos had shorter anterior/posterior axes as well as crooked or bent tails. (B) Phenotype analysis after pax2a staining at 24 hpf resulted in about 63% embryos with a mild or severe affected phenotype after Hoxb7a overexpression compared to 13% in injected controls. (C) Pax2a staining after microinjection of different concentrations of mRNA. From left to right severe, mild affected and wild type (WT) embryos at 24 hpf. WT zebrafish show expression in the hindbrain, hindbrain-midbrain boundary, neural tube, mesoderm, optic stalk, otic vesicle, and pronephric duct. Microinjection 62.5 μM mRNA, 125 μM mRNA and 250 μM mRNA resulted in respectively 48%, 71% and 61% malformed zebrafish. There was no correlation between mRNA dosage and severity of malformation.