Figures & data
Figure 1. Unsupervised hierarchical cluster analysis of the 1000 most variable probes between chordoma samples and nucleus pulposus samples.
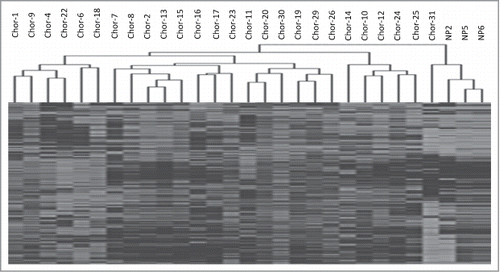
Figure 2. Genomic context of CpG methylation. (A) Differentially methylated probes in relation to gene. (B) Differentially methylated probes in relation to CGI context. (C) Sub-location of differentially hypermethylated probes in relation to gene. (D) Functional distribution of hypermethylated probes in relation to CGI.
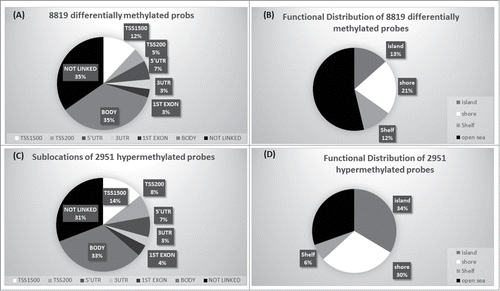
Table 1. Differentially methylated genes between recurrent and non-recurrent chordoma samples
Figure 3. Expression and methylation analysis in ES cell lines. Expression and methylation results for 3 selected genes in chordoma cell lines (PON3, SLC16A5, and NPR3; GAPDH as control). (A) Expression analysis using RT-PCR in 2 chordoma cell lines are shown for each gene with (-) indicating the samples without 5-azaDC treatment and (+) indicating the samples after 5-azaDC treatment. (B) COBRA results for the selected genes in cell lines. Undigested (U) samples were loaded next to the BstU1 digested samples (D). Two chordoma cell lines are shown for each gene and the positive control (Positive C) is in vitro methylated DNA. (*) indicates that the cell line is methylated. β-values from the HumanMethylation450 BeadChip array for UCH-1 cell line is also shown under each digest.
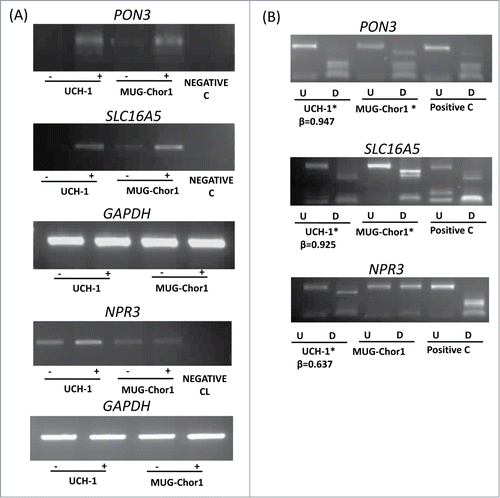
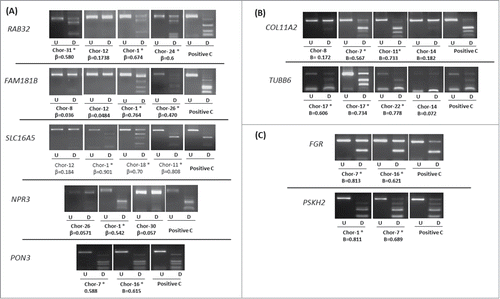
Figure 4. COBRA analysis for selected genes in chordoma samples. (A) Results of COBRA analysis for 5 genes (RAB32, FAM181B, SLC16A5, NPR3, and PON3) selected from differentially methylated gene list between recurrent and non-recurrent chordoma samples. (B) Results of COBRA analysis for 2 genes (COL11A2 and TUBB6) from differentially methylated probe list. (C) Results of COBRA analysis for 2 genes (FGR and PSKH2) selected from cancer-specific hypermethylated probe list. COBRA results are shown for chordoma samples with in vitro methylated DNA as positive control (Positive C). The digested samples (D) are loaded next to the undigested samples (U). (*) indicates methylated samples. β-value from the HumanMethylation450 BeadChip array for each selected sample is shown.