Figures & data
Table 1 Datasets used in the current study
Figure 1. Epigenomic similarity among the ENCODE cell lines. We computed the Spearman correlation coefficient between cell type-specific epigenomic enrichment profiles (Methods). The resulting correlation matrix was clustered using Euclidean/average clustering metrics, and visualized with darker blue/red gradient representing weaker/stronger epigenomic similarity, respectively. Each cell shows the numerical value of the corresponding Spearman correlation coefficient.
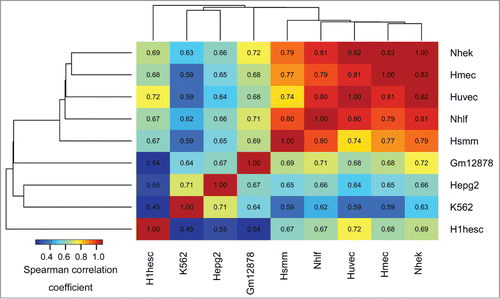
Figure 2. Epigenomic similarity among aDMRs and aGENs using the ENCODE data, H1hesc cell line. We computed the Spearman correlation coefficient between aDMR-and aGEN-specific epigenomic enrichment profiles (Methods). The resulting correlation matrix was clustered using Euclidean/average clustering metrics, and visualized with darker blue/red gradient representing weaker/stronger epigenomic similarity, respectively. Each cell shows the numerical value of the corresponding Spearman correlation coefficient.
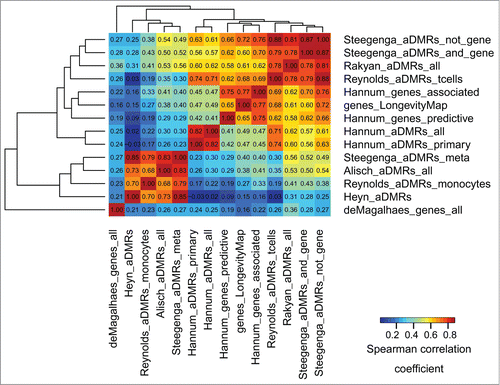
Figure 3. Histone modification marks (ENCODE) enriched/depleted in aDMRs and aGENs. Darker blue/red gradient highlights depleted/enriched associations, respectively. Red/green bar gradient defines frequency of epigenomic marks enriched/depleted in aDMRs and aGENs, respectively.
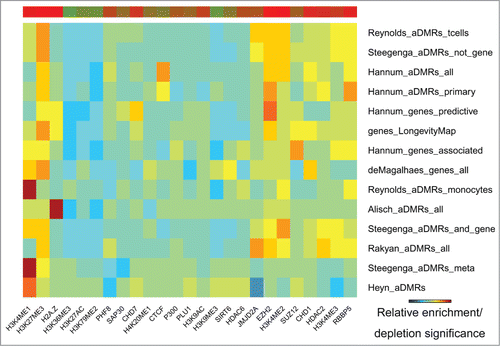
Figure 4. Transcription factor binding sites (ENCODE) enriched/depleted in aDMRs and aGENs. Darker blue/red gradient highlights depleted/enriched associations, respectively. Red/green bar gradient defines frequency of epigenomic marks enriched/depleted in aDMRs and aGENs, respectively.
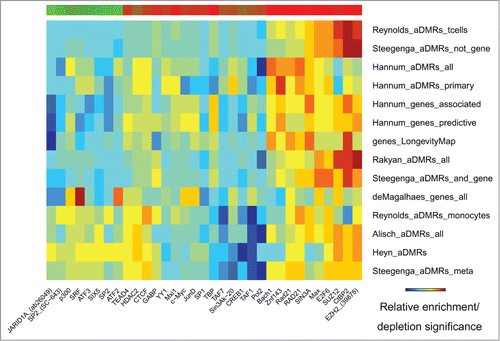
Figure 5. Chromatin states (ENCODE) enriched/depleted in aDMRs and aGENs. Darker blue/red gradient highlights depleted/enriched associations, respectively. Red/green bar gradient defines frequency of epigenomic marks enriched/depleted in aDMRs and aGENs, respectively.
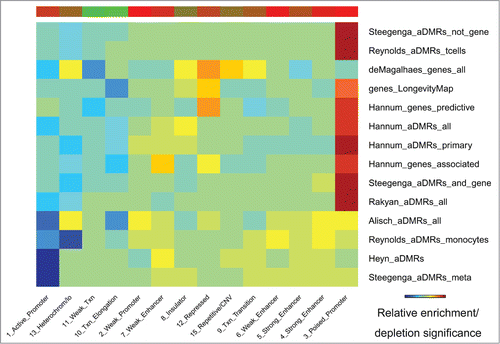
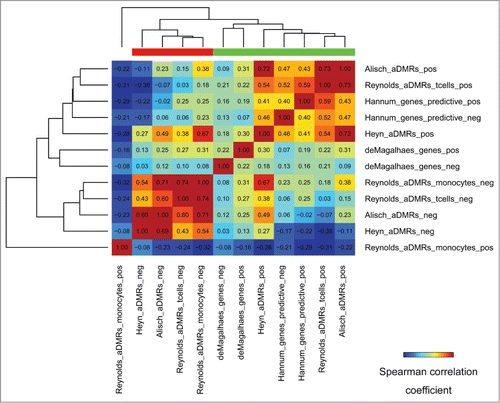
Figure 6. Epigenomic similarity among aDMRs hyper/hypomethylated with age, and aGENs showing increasing/decreasing gene expression with age (“pos/neg” postfixes). Pos/Neg aDMR-and aGEN epigenomic enrichment profiles obtained using H1hesc epigenomic data from the ENCODE project (see Methods) were correlated to each other using the Spearman correlation coefficient metric. The resulting correlation matrix was clustered using Euclidean/average clustering metrics, and visualized with darker blue/red gradient representing weaker/stronger epigenomic similarity, respectively. Each cell shows the numerical value of the corresponding Spearman correlation coefficient. Red/green bars define groups of aDMRs and aGENs showing high inter-group similarity.