Figures & data
Table 1. Characteristics of the study subjects by exposure groups
Figure 1. Flowchart of 450K Infinium Methylation BeadChip sample analyses. Note: *Differential methylation was defined as an |Δβ|>0.05 and false discovery rate-FDR<0.05.
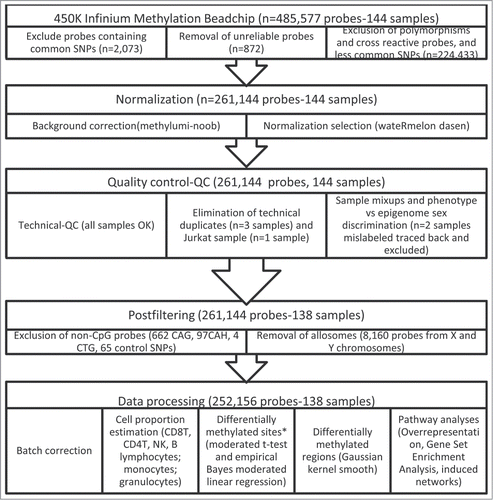
Figure 2. Heatmap of CpG sites with methylation levels associated with THM exposure (|Δβ|>0.05 and FDR<0.05). Note: DNA methylation heatmap of methylated genes passing FDR<0.05, in white blood cells DNA of persons exposed during lifetime to trihalomethanes. Each row represents a CpG site with columns representing each sample. The top dendrogram shows the results of an unsupervised hierarchical clustering of 138 samples based on 29 CpG sites, which separates those subjects flagged as highly exposed in average to lifetime THM levels >85 μg/L (marked as black in the bottom box), from those exposed to lower levels (the remainder columns). In the right box each site is marked to its corresponding region. A scatterplot of the actual lifetime THM levels is shown at the bottom box.
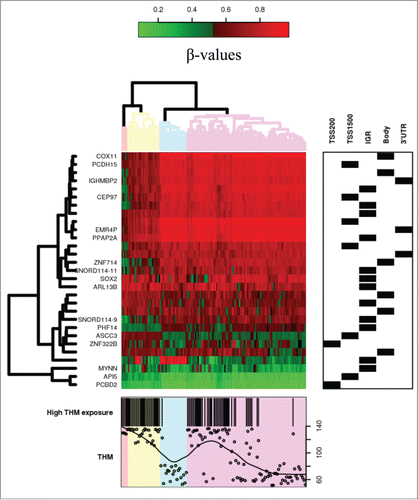
Table 2. Top CpG sites (n = 29) with methylation levels associated with THM exposure (|Δβ|>0.05 and FDR<0.05), after adjusting for covariables (age, sex, first principal component and blood cells proportion)
Figure 3. Network analysis of differentially methylated genes after adjustement by covariables. Expected Δβ is the methylation change expected after adjusting for age, sex, the first principal component and the estimated white blood cell proportion.
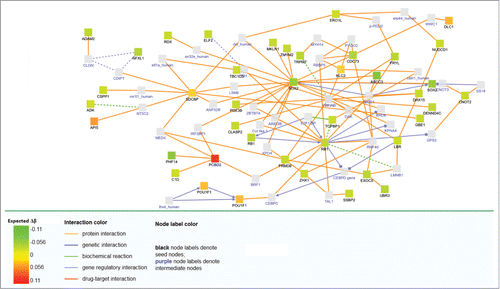
Table 3 Differentially methylated regions (n = 30) identified using a Gaussian kernel (DMRcate) with methylation levels associated with trihalomethane exposure ((|Δβ|>0.05 and FDR<0.05) after adjusting for covariables (age, sex, first principal component and blood cells proportion)
