Figures & data
Figure 1. TET1 overexpression and TET triple knockdown affect global 5hmC levels of HEK293 cells. (A) Quantitative real-time RT-PCR of induced vs. uninduced T-REx-293-TET1 cell lines demonstrates elevated TET1 mRNA levels after doxycycline induction of transgene expression. T-REx-293-GFP control cells (GFP) do not show elevated TET1 transcript levels upon induction. Measurements were performed in duplicates and presented as mean ± SD. (B) Western blot analysis of FLAG-tagged TET1 overexpression in T-REx-293-TET1 cell lines in induced and uninduced state with anti-FLAG (upper part) and anti-TET1 antibodies (lower part) shows inducible expression of TET1. (C) DNA dot blot analysis shows increased global 5hmC levels in induced compared to uninduced T-REx-293-TET1 cells. (D) Quantitative real-time RT-PCR demonstrates reduced TET transcript levels in T-REx-293-GFP cell lines treated with siRNAs against TET1, TET2 and TET3 when compared to scrambled controls. Measurements were performed in triplicates and presented as mean ± SD. (E) DNA dot blot analysis shows decreased global 5hmC levels in TET triple knockdown (TET kd) compared to scrambled control (scr) T-REx-293-GFP cell lines. TET1 #1–3: single-cell derived T-REx-293-TET1 cell lines; GFP #1–3: single-cell derived T-REx-293-GFP cell lines; +dox: doxycycline- induced transgene expression; -dox: uninduced control.
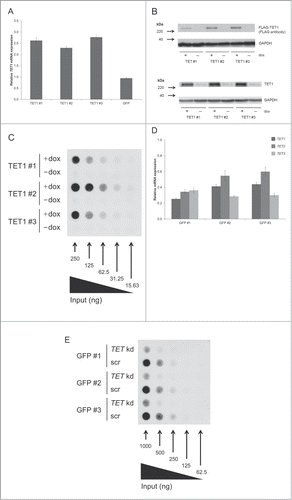
Figure 2. TET1 overexpression and TET triple knockdown affect DNA methylation levels of HEK293 cells. Shown are comparisons of mean methylation β values for different genomic regions after induced overexpression of TET1 (TET1 +dox; A–C) and triple knockdown of the TET enzymes (TET kd; D–F) compared to the respective uninduced (TET1 -dox) or scrambled controls (scr). The methylation status of more than 450,000 CpG sites was measured by Illumina Infinium HumanMethylation450 BeadChips and quantitatively analyzed on the region level for the 3 replicate cell lines of each experiment. The transparent blue areas correspond to point density with the 1% of the points in the sparsest populated plot regions drawn explicitly. The 1,000 best ranking differentially methylated promoters (A, D), gene bodies (B, E) and CGIs (C, F) are represented by red dots. ρ = Pearson correlation coefficient.
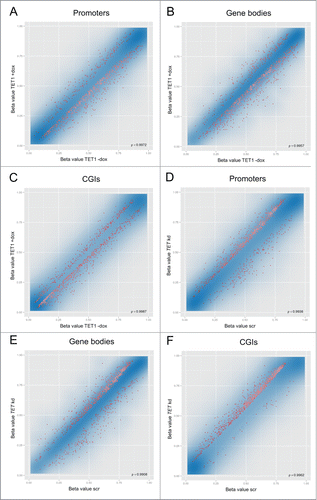
Figure 3. Reciprocal methylation changes of TET1 overexpression and TET triple knockdown. Shown are the distributions of the 500 top-ranked differentially methylated promoters, gene bodies, and CGIs toward hypo- (↓) and hyper-methylation (↑). In general, TET1 overexpression was associated with DNA hypomethylation while TET knockdown was associated with DNA hypermethylation (both in bold). Overlaps are shown in the right hand column.
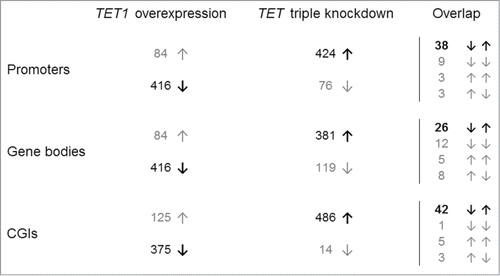
Figure 4. TET1 overexpression and TET triple knockdown affect promoter methylation levels depending on gene expression levels. The top-ranked differentially methylated gene promoters that showed a methylation decrease after TET1 overexpression (A) or a methylation increase after TET knockdown (B) are grouped according to gene expression quartiles of the respective control cells. Comparisons between expression and methylation were only performed for genes with identical gene symbol annotations in both data sets (TET1 overexpression: n = 234, TET triple knockdown: n = 322). Q1 – Q4: gene expression quartiles ranging from lowest to highest expression levels.
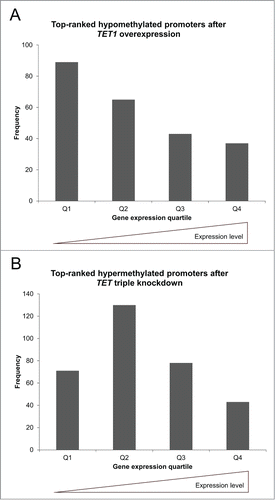
Figure 5. TET1 overexpression leads to increased 5hmC levels at most 5hmC sites. Two T-REx-293-TET1 cell lines in uninduced and induced state were subjected to RRHP. Shown are averaged read counts reflecting 5hmC abundance in TET1-overexpressing (TET1 +dox) and control cells (TET1 -dox). CpG sites with less than 5 reads were considered zero. The black line illustrates a perfect positive correlation. ρ = Pearson correlation coefficient.
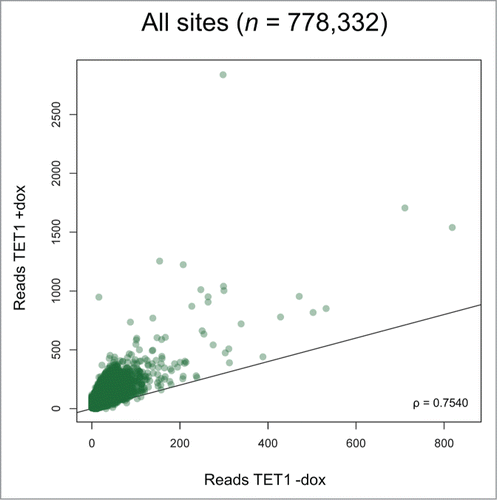
Figure 6. TET1 overexpression does not change the relative 5hmC distribution among genetic elements. Presented are 5hmC read distributions for different genetic elements and regions in control (TET1 -dox) and TET1-overexpressing cells (TET1 +dox). CpG sites with 5 or more reads were counted.
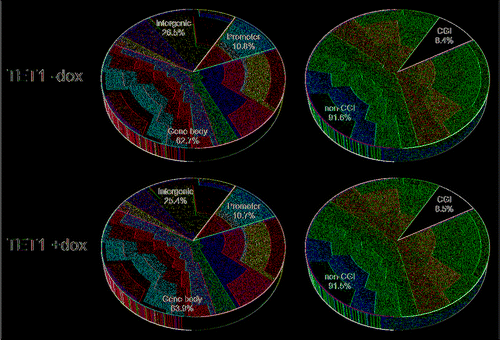
Figure 7. TET1-mediated 5mC oxidation is associated with endogenous 5hmC levels. Endogenous 5hmC read levels in control cells (reads TET1 -dox) are compared to the fold changes of 5hmC that occurred after TET1 overexpression (TET1 +dox). Sites with basal 5hmC levels corresponding to 15–20 reads showed the highest increase. Only CpG sites with 5 or more reads in TET1-overexpressing and control cells were considered. Boxes mark the interquartile range with whiskers indicating the 1.5 × interquartile range. Outliers beyond the whiskers are plotted.
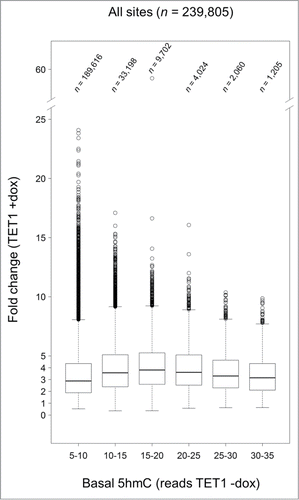
Figure 8. TET1 oxidates promoter 5mC depending on gene expression levels. The promoters showing the highest de novo or fold change increase of 5hmC (A) and those promoters characterized by a complete loss or lowest fold change of 5hmC upon TET1 overexpression (B) are grouped according to gene expression quartiles of uninduced control cells. Comparisons between expression and 5hmC methylation were only performed for genes with identical gene symbol annotations in both data sets (top de novo promoters: n = 372, top fold change promoters: n = 381, complete loss promoters: n = 195, lowest fold change promoters: n = 334). Q1 – Q4: gene expression quartiles ranging from lowest to highest expression levels.
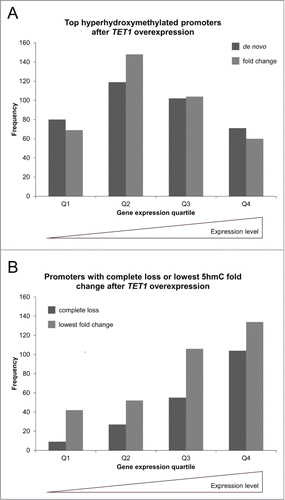
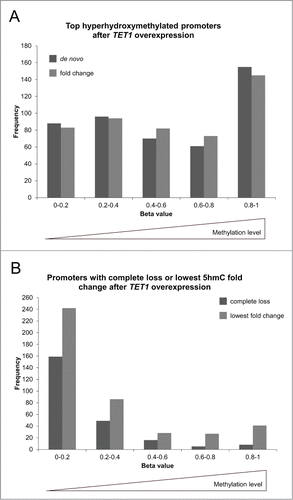
Figure 9. TET1 oxidates 5mC preferably at highly methylated promoters. The promoters showing the highest de novo or fold change increase of 5hmC (A) and those promoters characterized by a complete loss or lowest fold change of 5hmC upon TET1 overexpression (B) are grouped according to their basal, endogenous methylation levels as determined by the HumanMethylation450 BeadChip experiments. Comparisons between 5hmC and HumanMethylation450 data were only performed for genes with identical gene symbol annotations in both data sets (top de novo promoters: n = 470, top fold change promoters: n = 477, complete loss promoters: n = 237, lowest fold change promoters: n = 424).