Figures & data
Table 1. Characteristics of study population (n = 677) by birth weight category
Figure 1. Panel of genes identified to be differentially expressed by birth weight category (FDR < 0.05). Boxes represent the first to third quartiles of each distribution, with the median indicated by the horizontal bar. The upper whisker extends to the highest point within 1.5x inter-quartile range (IQR) from the upper quartile, and the lower whisker extends to the lowest point within 1.5× IQR from the lower quartile. Outlier data points are shown as filled circles.
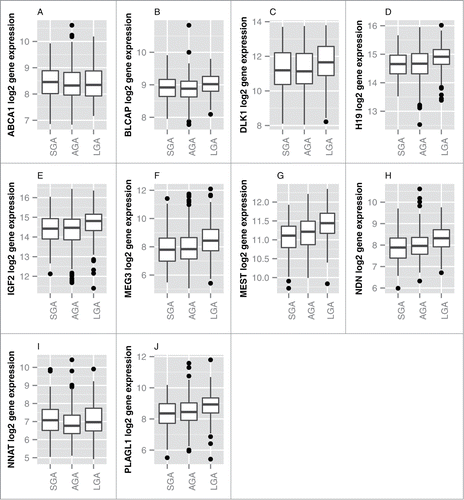
Figure 2. Odd ratios and 95% CI for SGA or LGA status for a log2 unit increase in gene expression. Multinomial regression analysis referenced against AGA status indicates that the expression levels of BLCAP, DLK1, H19, IGF2, MEG3, MEST, NDN, NNAT and PLAGL1 are positively associated with LGA status. NNAT is positively associated with SGA and MEST is negatively associated with SGA status. Odds ratios (OR) and 95% CI are shown on the x-axis for each gene's association with birth weight (y-axis).
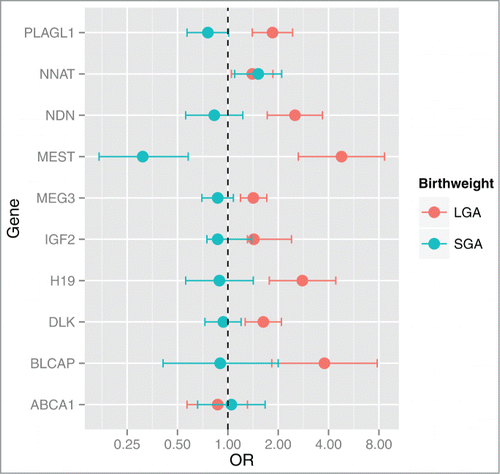
Figure 3. Correlation plot of expression of imprinted genes in the placenta. Pairwise correlations (Pearson R-values) are depicted for gene pairs labeled on the diagonal axes. Genes were ordered according to the factors they load onto, with factors indicated along the top axis. The side legend depicts the color gradient indicating directionality, positive (blue) and negative (red), and strength of the correlation (magnitude of correlation coefficient). Gray crosses in the plot indicate correlations with a significance level above 0.05.
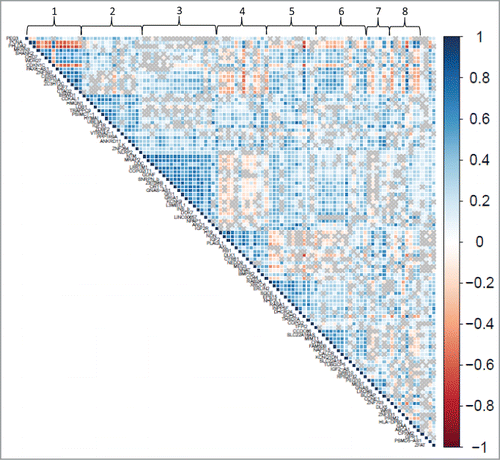
Table 2. Gene loadings of factors derived from factor analysisa
Figure 4. Odd ratios and 95% CI for SGA or LGA status for a log2 unit increase in factor scores. Multinomial regression analysis referenced against AGA status indicates that Factors 4 and 7 are positively associated with LGA status. Additionally, Factor 7 is negatively associated with SGA status and Factors 5 and 8 are negatively associated with LGA status. Odds ratios (OR) and 95% CI are shown on the x-axis for each factor's association with birth weight (y-axis).
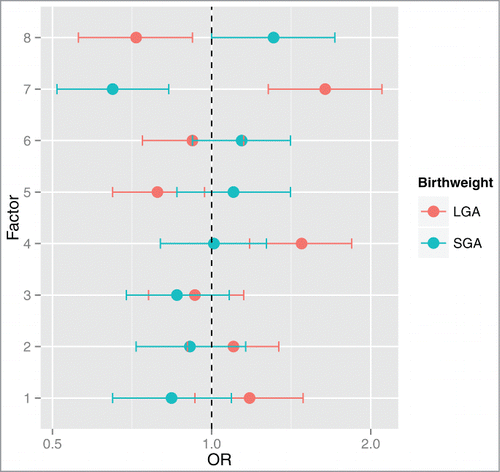
Figure 5. MEST contains differentially methylated sites based on birth weight status. Genomic coordinates of CpG loci profiled on the Illumina 450K array are provided on the x-axis and the extent of methylation (y-axis) is represented by the β value, with 1 indicating fully methylated and 0 indicating fully unmethylated. The transcription site is indicated by the dashed vertical line. Higher median MEST methylation levels are observed among SGA infants (red).
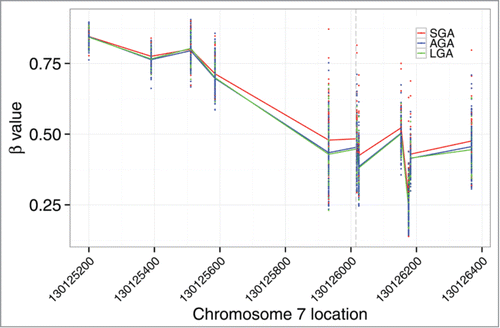
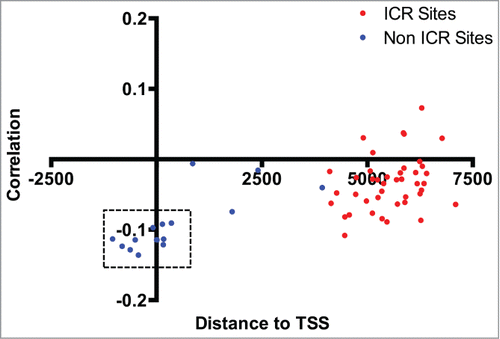
Figure 6. Correlation between MEST methylation and expression levels. Distance to the transcriptional start site (at 0) of the MEST gene (x-axis) and correlation coefficient (y-axis) for individual CpG loci profiled on the Illumina 450K array are shown, and sites in the known imprinting control region are designated as red dots. A modest correlation is observed between MEST expression level and methylation sites clustered around the transcription start site. CpG loci corresponding to sites differentially methylated by birth weight status are indicated by dashed rectangles.