Figures & data
Figure 1. Signatures of quantitative reduced representation bisulfite sequencing (Q-RRBS). (A) A schematic of the Q-RRBS method used for starting materials from either a trace amount of cells or single cells. Briefly, we first performed cell lysis, MspI digestion, repairing/A-tailing, UMI-adapter ligation, and bisulfite treatment in single-tube reactions, and then performed amplification, sequencing, and deduplicated analysis. Adapters containing UMIs (unique molecular identifiers) at their 3′ ends are represented by the short, variously colored boxes within the ellipse. The random-labeling step showed that both ends of each double-stranded DNA molecule randomly ligated with one of the 26 distinct UMI adapters. After deduplicated analysis, the 5 identifiers indicated that only 5 molecules were present, whereas the original analysis showed that 20 sequencing reads were present. (B) An example of single-molecular-fragment-derived (SFD) duplicates that contained identical UMIs (6-bp sequences located at both ends of black lines) and aligned to the same position of the genome. This type of SFD duplicates contained 6 fragments that displayed homogeneity of the DNA methylation pattern (same distribution of methylated CpGs in the sequencing reads). Filled circles represent methylated CpGs. (C) Of the 6,306,437 types of SFD duplicates that derived from 6,306,437 positions of the diploid genome, 98.3% displayed homogeneity of the DNA methylation pattern (shown by the example in ) in each type of the SFD duplicates.
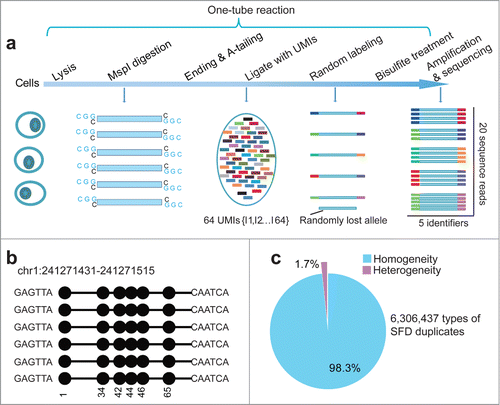
Figure 2. Duplication-induced artifacts in reduced representation bisulfite sequencing (RRBS) data. (A) Deviation of coverage-depth and methylation level between original and deduplicated analysis. In the original and deduplicated analyses, the total original reads and the deduplicated reads were used for subsequent analysis, respectively. The Circos plot on left displays the methylation level and depth of 5,246 CpG sites (differential methylation level > 0.2 between original analysis (blue) and deduplicated analysis (red; coverage ≥ 2). The right plot shows a representative locus (chr1:44,401,940-44,401,987) of significant deviation between the original and deduplicated analysis. (B) Distribution of CpG sites showing different DNA methylation levels in each set of original (blue dotted lines) and deduplicated (orange solid lines) data for the single-cell (SC), dozens-of-cells (DC), and 30 ng of MCF-7 DNA samples. The y-axis shows the number of counts in each range of methylation level. (C) Scatter plots of correlation coefficients between the methylation levels determined by means of deduplicated analysis and original analysis for each sample. We selected CpGs exhibiting methylation level ranging from 20% to 80%, as determined through deduplicated analysis. Horizontal and vertical axes represent the methylation levels of CpGs determined from the deduplicated analysis and original analysis, respectively. Color bars, ranging from pink to blue, represent the increase of CpG density. Correlation coefficients are shown above the scatter plots.
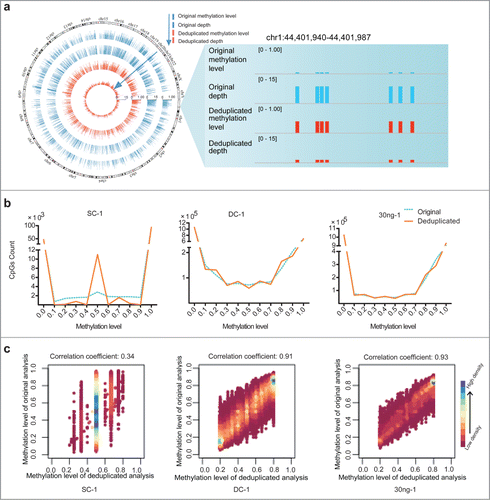
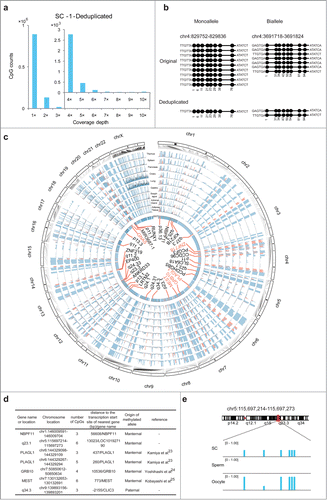
Figure 3. Analysis of quantitative reduced representation bisulfite sequencing (Q-RRBS) data obtained from HEK293T single-cell samples. (A) Bar plots of CpG counts at depths ranging from 1× to 10× after deduplicated analysis for the single-cell sample SC-1. The y-axis shows the counts of the CpGs corresponding to each distinct coverage depth. (B) Examples showing the ability of Q-RRBS to accurately identify the methylation pattern of monoalleles or bialleles for specific regions. The left plot shows that although 9 reads were aligned to the same locus of the genome from the original analysis, only the monoallele of this locus was detected from the deduplicated analysis. The right plot presents the results for certain biallelic loci that were detected by performing Q-RRBS with deduplicated analysis. (C) Circos plot of the methylation state of HEK293T cells and 9 differentiated tissues (thymus, spleen, pancreas, ovary, lung, gastric, esophagus, adrenal gland, and adipose tissues) at previously identified allele-specific DNA methylation (ASM) regions, which were obtained by using Q-RRBS and pooling the data for SC-1 and SC-2 samples. The methylation levels of CpGs from different tissues or HEK293T cells are depicted on different tracks. Red bars represent cell type-specific ASM regions that were intermediately methylated (methylation level: 0.25–0.75) in at least 5 of the 9 tissues. Here, 33 common or cell type-specific ASM regions are annotated in the core, in which the known or reported imprinted genes are shown in red. (D) Identification of maternal or paternal origin for 7 monoallelic methylation regions from the aforementioned 33 ASM regions. (E) Snapshot of methylation levels for a monoallelic methylation region (LOC101927190) in HEK293T cells, sperm, and oocytes.