Figures & data
Figure 1.(See previous page) DUX4 activation during myogenic differentiation coincides with SMCHD1 reduction. (A) Immunofluorescence microscopy analysis confirmed the typical DUX4 protein expression pattern in myosin positive, multinucleated myotubes derived from FSHD1 and FSHD2 individuals. Images were taken using a 200x magnification, scale bars are displayed. (B) Quantitative mRNA analysis in control (C), FSHD1 (F1), and FSHD2 (F2) primary myoblasts (B) and myotubes (T) showing increased levels of DUX4 expression upon differentiation. GAPDH and GUSB were used as reference genes, n indicates number of samples, error bars display SD, and significance was calculated using a 2-tailed Student t-test. (C) Quantitative mRNA analysis showed robust ZSCAN4 activation upon myogenic differentiation in FSHD myotubes. GAPDH and GUSB were used as reference genes, n indicates number of samples, error bars display SD, and significance was calculated using a 2-tailed Student t-test. (D) Western blot analysis showing reduced levels of SMCHD1 upon muscle cell differentiation in a primary muscle cell culture derived from a control individual. Myoblasts (B) were differentiated into myotubes (T) for 48 h. H3 serves as a control for equal protein loading, α-ACTIN serves as a control for the induction of myogenic differentiation. (E) Western blot analysis of a control derived primary fibroblast undergoing forced myogenesis by ectopic MyoD expression for 48, 72, or 94 h showed a decrease in SMCHD1 protein expression. TUBULIN serves as a loading control. (F) Duplicate RT-PCR analysis of DUX4 expression upon ectopic MyoD expression in a Control 1, FSHD1 1 or FSHD2 1 fibroblast (Table S2) revealed DUX4 activation in patient cells exclusively. Ectopic GFP expression was used as a control and GUSB serves as an internal PCR control. (G, H) Normalized ChIP qPCR analysis of SMCHD1 binding at D4Z4 in isogenic fibroblasts (F), myoblasts (B), and myotubes (T) derived from the same control individual (panel G) and 3 independent control derived myoblast – myotube pairs showed the highest SMCHD1 abundance in myoblasts with a further decrease during myogenesis. Error bars display SD in panel G and H and significance was calculated using a 2-tailed Student's test. NS = not significant; * = P <0.05; ** = P<0.005.
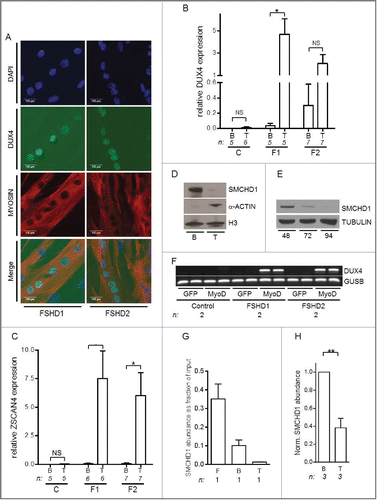
Figure 2. (See previous page) SMCHD1, but not SUV39H1 and Cohesin, regulates DUX4 expression. Western blot confirmation of (A) SMCHD1, (B) SUV39H1, (D) RAD21, and E) SMC3 knockdown in control myotubes expressing the indicated lentiviral transduced shRNAs. Tubulin was used as a loading control. Representative blots of at least duplicate experiments are shown. (C, F) Standard gel electrophoresis analysis of DUX4 expression upon lentiviral knockdown of SMCHD1, SUV39H1, RAD21 and SMC3 in control myotubes. Only depletion of SMCHD1 resulted in reproducible activation of DUX4 transcription. Representative gel photos are shown of at least duplicate experiments. The PCR fragment visible in one RAD21 knockdown was sequenced and shown to be the product of an a-specific amplification. (G) Western blot analysis confirms a 2-3 fold increase of SMCHD1 expression upon its lentiviral transduction in 2 FSHD1 and 1 FSHD2 myotube cultures. GFP transduced myotubes served as a negative control. Numbers indicate normalized relative expression levels of SMCHD1 using Tubulin as a loading control followed by setting normalized SMCHD1 levels of GFP transduced samples (only expressing endogenous SMCHD1) to 1. (H) Expression levels of DUX4 were significantly reduced upon ectopic expression of SMCHD1 in 2 FSHD1 and 2 FSHD2 myotube cultures. Relative DUX4 expression was calculated for each sample by normalization to GUSB and GAPDH housekeeping genes. Bars show values of each samples adjusted to the expression value of GFP transduced sample as 1. (I) Expression levels of the DUX4 target genes RFPL2, ZSCAN4, and TRIM43 showed a significant reduction upon ectopic expression of SMCHD1 in 2 FSHD1 and 2 FSHD2 myotubes, concordant with decreased DUX4 protein expression. Expression levels were normalized as described for panel Figure 1A. For panel H, I: Error bars display SD and significance was calculated using a 2-tailed Student's t-test. All P <0.0005 indicated by ***.
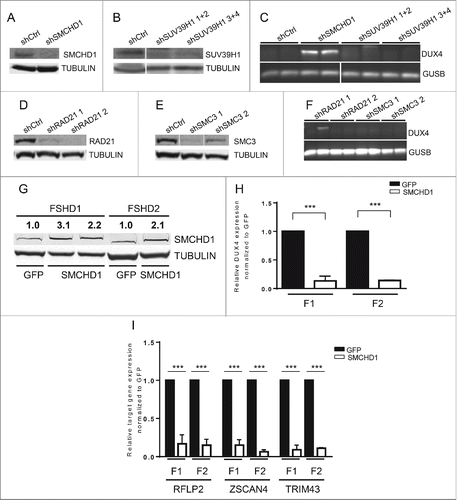
Figure 3. SMCHD1 depletion at D4Z4 leads to increased H3K27me3 and PRC2 binding. (A) ChIP-qPCR analysis showed a statistically significant reduction of SMCHD1 at qD4Z4 upon its lentiviral knockdown in control myotubes with 2 independent shRNA constructs in 2 independent control myotube cultures. (B) ChIP-qPCR analysis of H3K27me3 at qD4Z4 upon SMCHD1 depletion showed a significant increase at qD4Z4. Enrichment values were normalized to H3 enrichment values. (C) Normalized ChIP-qPCR analysis of SUZ12 upon SMCHD1 depletion showed a statistically significant increase at qD4Z4 in 2 independent experiments performed on 2 control myoblast cultures. n indicates sample size, error bars indicate SD, and significance was tested with Student t-test. (D) ChIP-qPCR analysis of H3K27me3 at qD4Z4 showed a significant increase in FSHD2 myotubes compared to controls. Enrichment values were normalized to H3 enrichment values. (E) ChIP-qPCR analysis of SUZ12abundance showed a significant increase in FSHD2 myotubes at qD4Z4. On panel A, B, D, and E, n indicates sample size, error bars display SD, and significance was tested using a one way-ANOVA followed by Bonferroni multiple comparison test. *= P <0.05 **= P<0.005 ***= P<0.0005.
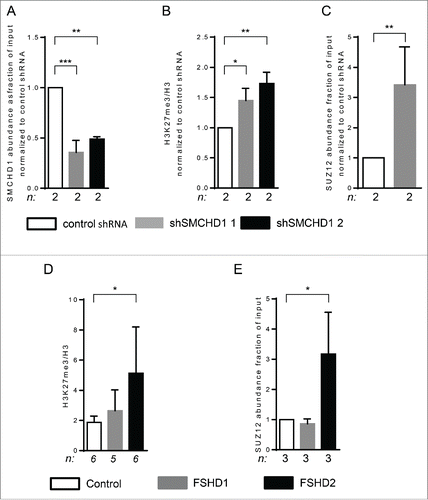
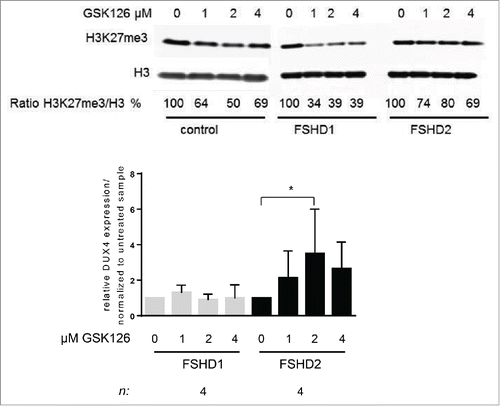
Figure 4. Treatment of FSHD2 myotube cultures with EZH2 inhibitor GSK126 increases DUX4 levels. (A) Western blot analysis of control, FSHD1, and FSHD2 myotube samples after treatment with 0, 1, 2, and 4 μM GSK126. Blots were probed with H3K27me3 antibody and H3 antibody as loading control. Shown are the reduced ratios of H3K27me3:H3 signal intensities in samples treated with GSK126 compared to the untreated sample as a result of EZH2 inhibition. (B) qRT-PCR analysis of DUX4 expression in FSHD1 and FSHD2 myotubes after GSK126 treatment shows that relative DUX4 transcript levels are significantly increased in FSHD2 samples treated with 2 μM GSK126 but not in FSHD1 myotubes. Graph shows the results of 4 FSHD1 and 4 FSHD2 cell lines. Results of control cell lines are not shown, DUX4 transcript was not detectable. Error bars show SD, n indicates number of independent cell lines, and significance was tested by 2-way ANOVA followed by Bonferroni multiple comparison test. *=P <0.05.