Figures & data
Table 1. Top 15 ASM sites in whole blood, ASM score averaged across individuals.
Table 2. Top 15 ASM sites in cerebellum, ASM score averaged across individuals.
Table 3. Top 15 ASM sites in cortex (BA9), ASM score averaged across individuals.
Figure 1. Allelic-skewing is less prevalent and less variable across cortical regions compared to cerebellum and whole blood. (A) The average proportion of informative amplicons showing an ASM score ≥ 0.10 for the 6 cortical regions profiled as well as averaged across the cortical areas (Ctx) is consistently lower (ASM score range = 0.34–0.61%) than in cerebellum (Cer) (ASM prevalence = 1.14%) or blood (ASM prevalence = 0.84%). Standard errors are shown for tissues for which samples were available from all 3 individuals. (B) – (E) Correlations of ASM scores are shown with each point representing one probe in one individual. Probes classified as allelically-skewed at an ASM score ≥ 0.10 in only one of the 2 compared tissues are highlighted in red. A higher degree of between-tissue variability is observed between cerebellum, cortex, and whole blood than between different cortical regions (shown as an example is BA8 vs. BA10). This difference becomes even more pronounced when restricting the set of probes to those that show allelic-skewing at an ASM score ≥ 0.10 in at least one of the 2 compared tissues (see subset correlation r’).
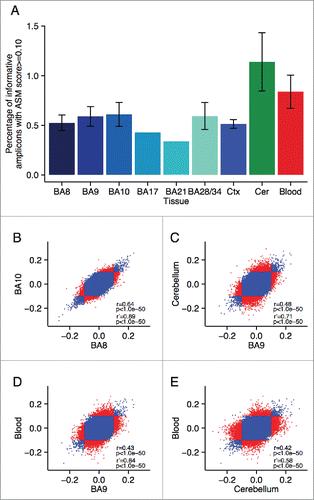
Figure 2. Multiple loci are characterized by consistent allelic-skewing of DNA methylation across all tissues. Heatmaps display allele signal intensities for genomic DNA (G), MSRE-digested DNA (D) and fully unmethylated, MSRE-digested DNA (U) in all tissues. A and B denote the 2 alleles of the SNP and brightness represents the quantile normalized signal intensity, with the scale displayed below the heatmap. Shown are the 2 top-ranked probes characterized by consistent ASM across tissues. These two probes were informative in (A) individual 2 for rs10234308 and (B) individual 3 for rs12493005.
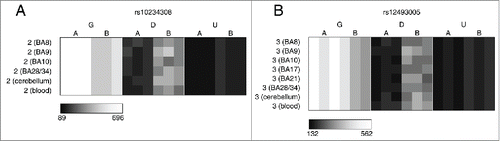
Figure 3. A number of loci are characterized by allelic-skewing of DNA methylation in only one tissue with minimal ASM present in any of the other tissues examined. Heatmaps display allele signal intensities for genomic DNA (G), MSRE-digested DNA (D) and fully unmethylated, MSRE-digested DNA (U) in all tissues. A and B denote the 2 alleles of the SNP and brightness represents the quantile normalized signal intensity, with the scale displayed below the heatmap. Two of the top-ranked cerebellum-specific ASM signals are (A) rs959246 (informative for individual 3) and (B) rs2252267 (informative for individual 2). The tissue-specific patterns of DNA methylation in these 2 loci were validated by clonal bisulfite sequencing, confirming the findings from the MSNP assay (C,D). Each row represents a single DNA molecule, with black dots depicting methylated cytosines and white dots depicting unmethylated cytosines. The percentage of methylated cytosines for each sample is displayed below the plots. The amplicon spanning the DMR associated with rs959246 in (C) did not encompass a SNP enabling us to distinguish between the 2 alleles, however the methylation pattern shows evidence for tissue-specific intermediate methylation (IM) in the cerebellum. G and A in (D) denote the 2 alleles determined by SNP variation within the amplicon in heterozygous individual 2. Cerebellum-specific hypomethylation of the A allele is observed surrounding rs2252267, with individual 3 (homozygous for the G allele) being highly methylated in all 3 tissues.
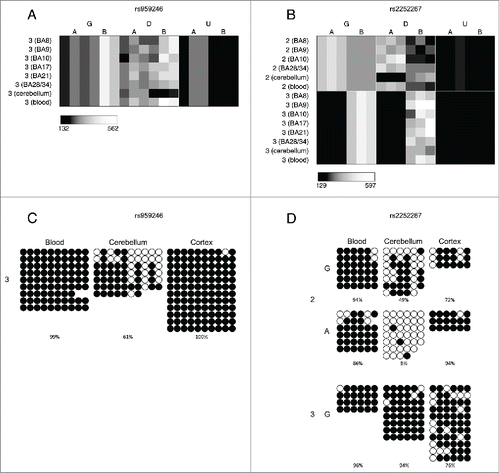
Table 4. Top 15 loci characterized by consistent ASM across cerebellum, whole blood, and cortex (BA9).
Table 5. Top 15 tissue-specific ASM sites, defined by highly variable ASM scores across cerebellum, whole blood, and cortex (BA9).
Figure 4. Cases of allelic-flipping of DNA methylation between individuals are found both in known imprinted gene clusters as well as regions not previously confirmed as being imprinted. The heatmaps show allele signal intensities for genomic DNA (G), MSRE-digested DNA (D) and fully unmethylated, MSRE-digested DNA (U) in all tissues. A and B denote the 2 alleles of the SNP and brightness represents the quantile normalized signal intensity, with the scale displayed below the heatmap. (A)-(C) Three probes in the vicinity of the known imprinted cluster on chromosome 15q11.2 show variable ASM and allelic-flipping in cerebellum [(A) rs940596, (B) rs11633486, (C) rs11854691]. (D) Allelic-flipping of DNA methylation across all tissues is observed in the vicinity of WRB (rs2244352), which has not been reported as imprinted previously.
![Figure 4. Cases of allelic-flipping of DNA methylation between individuals are found both in known imprinted gene clusters as well as regions not previously confirmed as being imprinted. The heatmaps show allele signal intensities for genomic DNA (G), MSRE-digested DNA (D) and fully unmethylated, MSRE-digested DNA (U) in all tissues. A and B denote the 2 alleles of the SNP and brightness represents the quantile normalized signal intensity, with the scale displayed below the heatmap. (A)-(C) Three probes in the vicinity of the known imprinted cluster on chromosome 15q11.2 show variable ASM and allelic-flipping in cerebellum [(A) rs940596, (B) rs11633486, (C) rs11854691]. (D) Allelic-flipping of DNA methylation across all tissues is observed in the vicinity of WRB (rs2244352), which has not been reported as imprinted previously.](/cms/asset/dacac8a2-1e6b-4385-a673-c4a04cd04437/kepi_a_1127479_f0004_b.gif)
Table 6. Top 15 variable ASM sites, defined by average range of ASM scores between individuals across cerebellum, whole blood, and cortex (BA9).
Figure 5. Regions around variable ASM sites are enriched for genomic domains characterized by intermediate DNA methylation. Flanking regions (1 kb) of variable ASM sites show a significant enrichment in intermediately methylated probes on the Illumina 450K Human Methylation Array (P range = 6.82 × 10−11 – 0.005). The scatter plots (A)-(D) show the location of genes and imprinting control regions (ICR) if overlapping the plotting window, as well as the location of the SNP from the MSNP assay (gray vertical line). These intermediate DNA methylation patterns span several known imprinted regions, for example (A) a flanking region of rs220030 in the SNRPN imprinted DMR, and (B) a region around rs2735971 in the imprinted gene H19. (C) Other sites showing allelic-flipping and intermediately methylated flanking regions lie in areas not previously known to be imprinted, for example rs2244352, which lies in an intron of WRB. (D) A polymorphic ASM pattern is observed in a flanking region around rs2346019, a downstream gene variant of VTRNA2-1, in which the majority of samples display intermediate DNA methylation in cortex (BA9) and whole blood. Of note, 4 samples show consistent hypomethylation across this region.
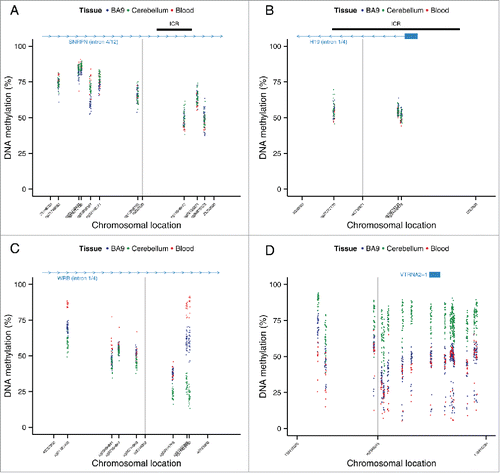
Figure 6. Regions around variable ASM are enriched in intermediate DNA methylation. The distributions of DNA methylation at the 65 Illumina 450K Human Methylation Array probes within 1 kb of the top 100 variable ASM sites (Table S14) show an enrichment in intermediately methylated probes compared to DNA methylation levels across the whole array (shown in gray) in (A) cortex (BA9), (B) cerebellum, and (C) whole blood).
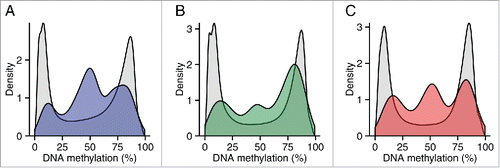
Table 7. Enrichment in intermediate methylation (IM) near ASM regions.
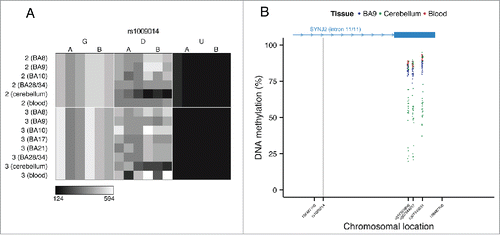
Figure 7. Genotype-driven ASM can be tissue-specific. (A) The tissue-specific ASM site rs1009014, located in an intron of SYNJ2, shows allelic-skewing of DNA methylation in cerebellum (ASM score = 0.16) but not in cortex (ASM score = 0.04) or whole blood (ASM score = 0.03). (B) DNA methylation levels for Illumina 450K Human Methylation Array probes in cerebellum across a flanking region of this locus exhibit a genotype-driven tissue-specific DNA methylation pattern. The scatter plot shows the location of SYNJ2 (transcript variant 1), as well as the location of the informative SNP from the MSNP assay (gray vertical line).